2D points 3D Objects 3D Points Boundary Cursor position Data for selected object Domain Expansion Field Field components Field formula Field tubes Function Generate expansions along 2D boundary Generate expansions for 3D objects Grid formula Grid transformation Info and movie directives Insert Integral Modify 2D expansions MMP Movie Open GL window PET basis PFD (predefined FD) Project Space, plane, arrow, or point Tools and draw Transformation data Window
Press the MMP button
![]() or
select MMP... from the Tools dialog to
open this dialog.
or
select MMP... from the Tools dialog to
open this dialog.
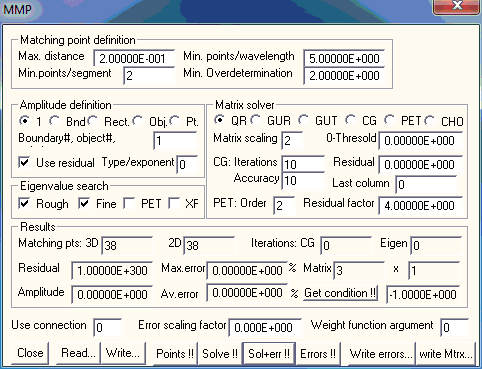
The MMP dialog allows you to set the MMP parameters and to run MMP for solving a given model. Note that an MMP model consists of the data specified in the Project, Boundary, Domain, and Expansion dialogs. When the MMP amplitude is defined by an integral the data in the Integral dialog is also required.
The MMP matrix is computed from the boundary conditions imposed in the matching points on the boundaries. Since the boundaries themselves contain no information on the matching point distribution, MMP computes the matching points upon request, using the parameters specified in the Matching point distribution group.
Select the maximum distance allowed between neighboring matching points in the Max. distance box. You can select a value larger than the total length of all boundaries, when you want to make this parameter ineffective.
For high frequency applications, the wavelength plays an important role. Useful models can only be obtained when the distance between neighboring matching points is considerably smaller than the shortest wavelength in a boundaries corresponding domains. You can define the minimum number of matching points per wavelength in the Min. points/wavelength box. This parameter becomes ineffective if it is less than 1. Reasonable values are in the range 4 to 20.
The boundaries consist of segments that are arcs, straight lines or spline sections. If the maximum distance between neighboring matching points defined in the Max. distance box and the distance computed from the Min. points/wavelength box is longer than the length of a segment, it may not be assigned a matching point. Consequently, the segment would be ignored by MMP. This can be avoided by setting the minimum number of points per segment in the Min.points/segment box greater than 0.
Multipoles and Bessel expansions are the most important MMP expansions. For these expansions, the angle between two neighboring matching points seen from the origin of the expansion should not exceed a certain value that depends on the maximum order of the expansion. From this, another restriction results that has an influence on the matching point distribution. When the parameter in the Min. Overdetermination box is equal to or greater than one, MMP will insert an additional matching point when the ‘view angle criterion’ is violated. This parameter increases the overdetermination of the MMP matrix. The default value 2 is reasonable in most cases. Higher values cause longer computation time and increase the reliability of the results.
For eigenvalue problems, the definition of the amplitude plays an important role. Usually, it will be defined either by a boundary integral or by integrating over a rectangular area. OpenMaXwell also contains integrals over the surface of 3D objects. These two(three) types of integrals are defined in the Integral dialog. You must specify whether you want to use boundary or rectangular integrals in the corresponding Bound. and Rect. (and Obj.) boxes. When you select Bound. (or Obj.) you must also specify the corresponding boundary (or 3D object) number in the Boundary (Boundary or object #) box. If the boundary (or object) number is 0, the integrals over all boundaries (or 3D objects) are summed up.
If either the Bound. or the Rect. (or the Obj.) box is checked, the parameters of the MMP expansions are scaled in such a way that the corresponding integral will evaluate to one.
OpenMaXwell also provides the Pt. option for advanced tests. It uses a test point located at the origin of the coordinate system defined for spherical area integrals (open the Integral dialog and press the 3D location button). In this test point, the complex field F defined in the integral dialog is evaluated and the inverse of the following 9 components are used as eigenvalue search functions: Abs(Fx), Abs(Fy), Abs(Fz), Re(Fx), Im(Fx), Re(Fy), Im(Fy), Re(Fz), Im(Fz). For the eigenvalue search, the number of the search function to be used is specified in the Type/exponent box, but this usually does not lead to good results. When you save the rough search data on a FLD file (check the FLD file box in the Project dialog), you may plot the 9 search functions as follows. 1: Re(Ex), 2: Re(Ey), 3: Re(Ez), 4: Re(Hx), 5: Im(Hx), 6: Re(Hy), 7: Im(Hy), 8: Re(Hz), 9: Im(Hz). Note that OpenMaXwell plots, for example, Im(Hy) when you select the y component of H field, turn Average and Phi off and specify 90 degree phase angle in the Derived Field group of the Field dialog.
For non-eigenvalue problems, the amplitude is often given by the incident field, i.e., the excitation. When you want the amplitude of the excitation equal to 1 but don’t want the integral to be evaluated, select the 1 box. This setting is also useful for simple eigenvalue problems.
Eigenvalue problems such as guided waves and resonators require a search for the eigenvalues. This search is done in two steps. Usually, a rough search over the search space defined in the Project dialog is followed by a fine search that is terminated as soon as the criteria defined in the Project dialog are met. This is done when both the Rough and Fine boxes of the Eigen search (or Eigenvalue search) group are checked. When you think that you already have entered a sufficiently good eigenvalue guess, you can skip the rough search. When you would like to inspect the search space before you start the fine search, you can turn the fine search off.
The most primitive eigenvalue solver of OpenMaXwell usually searches for the minims of the function Residual/Amplitude. In this eigenvalue solver the last expansion plays the role of the excitation. Therefore, the proper selection of the last expansion in the Expansion dialog may be important for the correct solution of tricky eigenvalue problems. An alternative eigenvalue solver is used in the Method of Auxiliary Sources (MAS). This eigenvalue solver introduces an extra excitation that usually is either an auxiliary monopole or an auxiliary multipole with fixed order. The auxiliary excitation is defined in the Expansion dialog and used during the eigenvalue search, but it is excluded when the eigenfield is evaluated (For doing this, set an appropriate value in the use expansion box of the Field formula dialog. For example, when you have defined 17 expansions, use expansion –16 will exclude expansion 17 when the field is computed.). This method has not yet been fully analyzed, but it seems to work well especially for computing band diagram of a photonic crystal. The eigenvalues of the MAS method are located where the Amplitude and the Residual have their maxims, i.e., where the functions 1/Amplitude and 1/Residual have minims. When you want that OpenMaXwell searches for the minims of 1/Amplitude (instead of Residual/Amplitude) make sure that the Use residual box is not checked and that the Amplitude is properly defined in the Integral dialog. Note that best results were obtained for band structure computations of photonic crystals when the time average of the total energy density defined in a single test point (Integral over a rectangular area with an 1 by 1 grid) was used. It has been found by Esteban Moreno that the function 1/Amplitude may have two minims (“twin minims” instead of a single minimum. The two minims are very close to each other and separated by a sharp peak. At the location of this sharp peak, the Residual also has a sharp peak. Therefore, one can also search for the minims of the function 1/Residual. In order to do this, make sure that the Use residual box is checked and specify the value –1 in the Type/exponent box. Because of the sharpness of the peaks of the Residual, there is a high risk that the rough search routine does not detect all eigenvalues when the search grid is not extremely fine. Therefore, 1/Residual should only be used when a very accurate fine search is required. When one works with the 1/Amplitude search function, one should be aware of the “twin minims” that correspond to one single eigenvalue. The Use residual and the Type/exponent boxes allow one to specify rather different eigenvalue search functions that may be more appropriate in difficult situations. The search function is defined as follows: 1) When Use residual is checked, the search function is (Residual/Amplitude)**(1/Exponent). 2) When Use residual is not checked, the search function is 1/Amplitude for Exponent=0 and (1/Amplitude)**(1/Exponent). 2)/Residual**Exponent otherwise. Note that the Residual has an influence on the search function when Exponent is not zero even when Use residual is not checked. The methods with Use residual not checked and Type/exponentsufficiently big allow one to define a search function without “twin minims” and with less sharp peaks than the 1/Residual function. However, this method provides the risk of numeric underflow, especially when Type/exponentis too high. Note that you even can define the search function in such a way that the minims of the search function do not correspond to eigenvalues (Namely when the Exponent is negative). Therefore, the proper definition of the value in the Type/exponentbox requires some experience. When you are not sure, start with Type/exponent 0.
In order to overcome the eigenvalue search problems associated with the "twin minims", the current fine search routine can observe not only the search function mentioned above, when the value in the Type/exponent box is different from zero. It also can observe the alternative search functions 1/residual and 1/amplitude. This makes the fine search more robust for bandgap evaluations. This advanced procedure is applied when you check the XF box in the Eigenvalue search group. Note that "twin minims" may be avoided when small losses are introduced.This feature is under "construction".
More sophisticated eigenvalue search routines are based on mismatching error evaluations along all boundaries. These were implemented because of severe difficulties in the evaluation of flat band modes in metallic photonic crystals (lossy and dispersive). In order to use them, the Use residual box must be checked and the Type/exponent value must be bigger than 2. The following 9 quantities are defined as eigenvalue search functions:
residual / amplitude,
residual (Note that this value was 1/residual before the 2015 version!),
1 / amplitude,
relative error average,
relative error maximum,
absolute error average / field average,
absolute error maximum / field average,
absolute error average / field maximum,
absolute error maximum / field maximum.
The number of the search function to be used for the eigenvalue search is specified in the Type/exponent box. Since this must be bigger than 2, you cannot select type 1 or 2 when you want that OpenMaXwell computes all 9 search functions. When you select type 1 or 2, the old eigenvalue search routines are used - which are indeed based on the residual/amplitude evaluation. Usually, type 4, 6, 7 provide good results, the other types provide additional information that is valuable for sophisticated users. When you save the rough search data on a FLD file (check the FLD file box in the Project dialog), you may plot the 9 search functions as follows. 1: Re(Ex), 2: Re(Ey), 3: Re(Ez), 4: Re(Hx), 5: Im(Hx), 6: Re(Hy), 7: Im(Hy), 8: Re(Hz), 9: Im(Hz). Note that OpenMaXwell plots, for example, Im(Hy) when you select the y component of H field, turn Average and Phi off and specify 90 degree phase angle in the Derived Field group of the Field dialog.
The eigenvalue estimation technique is a special form of the PET that can be applied when the dependence of eigenvalues of a variable is computed. In this case, the parameter set to be estimated consists of the complex eigenvalue and of an internal parameter of the fine search. Check the PET box in the Eigen search (or Eigenvalue search) group to turn this type of PET on. Note that the PET and CG matrix solvers cannot be used when the eigenvalue estimation technique is applied.
For example, if you want to compute the frequency dependence of the propagation constant of a guided wave, OpenMaXwell can compute the propagation constant of the first frequency with both a rough and a fine search. The result may be a good estimate for the computation of the second frequency. Therefore, the rough search can be skipped for the remaining frequencies. When n+1 previous eigenvalues are known, these values can be used for obtaining an n-th order eigenvalue estimation of the current value.
You can select six different matrix solver techniques. 1) QR: QR decomposition of the Rectangular MMP matrix, 2) GUR: Givens Updating with a Rectangular MMP matrix, 3) GUT: Givens Updating on a Triangular matrix, 4) CG: Conjugate Gradients solution on the rectangular MMP matrix, 5) PET: Parameter Estimation Technique, and 6) CHO: Cholesky decomposition of the symmetric matrix S=M*M, where M is the rectangulat MMP matrix. M* is the adjoint of M. QR is currently by far the fastest solver of the rectangular MMP matrix. GUT and CHO do not explicitly store the rectangular MMP matrix and work directly on a triangular matrix that is considerably smaller than the rectangular one. Therefore, GUT and GUR are appropriate when you are short on memory. CHO is a bit faster than GUT, but it is less accurate and may even fail when the condition number of M is high. Therefore, CHO should only be used for testing. GUR is similar to GUT, but by setting up the rectangular MMP matrix first, it allows OpenMaXwell to introduce scaling and tests that can be helpful. CG is an iterative matrix solver. Therefore, you need to specify the maximum number of CG iterations to be performed. Moreover, you can define two additional stopping criteria for CG: 1) The Residual and 2) the Accuracy. CG will stop when either the reisual is smaller than the value defined in the Residual box or when the relative difference of the current and previous residual is less than 10-Accuracy, i.e., the value defined in the Accuracy box essentially denotes the accuracy in digits. Since the CG iterations may exhibit a staircase convergence, you might obtain inaccurate results when Accuracy is too small, for example, 1 or 2. When Accuracy is bigger than 14, this stopping criterion will never be met. Note that the number of CG iterations should be considerably smaller than the number of MMP parameters, i.e., of unknowns. Otherwise, CG might be more time-consuming than GUR and GUT. To obtain accurate CG results within a few iterations, the condition number of the MMP matrix should be not too high. OpenMaXwell evaluates the condition number with a singular value decomposition when you press the Get condition!! button. Before you press this button, the rectangular MMP matrix must have been compute either with GUR or CG. Note that the evaluation of the condition number is more time consuming than the evaluation of the matrix. For obtaining few CG iterations, OpenMaXwell should have a good starting value, i.e., a good estimate of the MMP parameters. Such a guess can be obtained from previous computation of slightly modified MMP models with the same number of unknowns. Usually, CG will start with the MMP parameters defined in the Expansion dialog. The most useful technique for MMP computations of a sequences of similar MMP models with a fixed number of unknowns is the PET. This technique starts with GUR for the first model and uses the results of one or several previous models for estimating the parameters for the subsequent models that will be solved with CG. The CG will then be stopped when the residual is less than Residual factor times the previous GUR residual. When this stopping criterion is not met before the maximum of CG iterations is reached, OpenMaXwell will restart GUR for finding the best solution. To avoid this, the Residual factor should be sufficiently high, but not too high (otherwise the CG results will be inaccurate. The PET order can be selected in the corresponding box. Note that the parameters of n+1 previous models must be stored for n-th order PET. Advanced OpenMaXwell users may optimize the basis functions used for the PET. By default, a power series extrapolation is used. It is not reasonable to use higher orders than 2 in this case.
When solving eigenvalue problems, the last expansion, i.e., the last column of the MMP matrix is crucial. Instead of reordering the expansions for pushing a certain column to the last position, you may define a non-zero value L in the Last column box. If this parameter is smaller than 1 or bigger than the number of columns of the MMP matrix, nothing happens. otherwise, the last column M is exchanged with the column M-L before the matrix solver is called. Then, the column M-L plays the role of the excitation.
Usually, the value of the Connect. (or Use connection) box is zero and all boundaries and expansions have the connection flag zero. When you use connections, some of the expansions and eventually also some of the boundaries have a different connection flag. In this case, MMP computes the MMP parameters of all expansions that are contained in the connection selected in the Connect. (or Use connection) box and it uses the boundary conditions on all boundaries that belong to the connection selected in the Connect. (or Use connection) box. A typical example for MAXaax.EXE is the near-to-far field transformation, where the near field is first computed in one connection and the far field is computed afterwards in another connection.
OpenMaXwell displays the results of the MMP computation in the corresponding group of the dialog. Note that you cannot immediately read the output in the Results group because the dialog closes when you start any MMP evaluation. Open this dialog again to read 1) the number of CG iterations that were performed (provided that CG was used), 2) the number of Matching points that were generated, 3) the value of the Residual and of the Amplitude (provided that the MMP matrix equation was solved), 4) the number of Eigen iterations that were performed (provided that an eigenvalue problem was solved an the Fine search was turned on), 5) the mismatching detected on the boundaries, i.e., its maximum and average relative errors (provided that the errors have been computed), 6) the number of rows and columns of the MMP matrix.
OpenMaXwell also outputs the size of the MMP matrix in the Matrix and x boxes. Moreover, the numbers of the 3D and 2D matching points are displayed in the boxes 3D and 2D respectively. The number of CG iterations and the number of eigenvalue iterations are displayed in the CG and Eigen boxes. As soon as the error distribution has been computed, the maximum and average values are displayed in the Max.error and Av.error boxes.
When you have computed the mismatch errors along the boundaries, OpenMaXwell draws lines perpendicular to the boundary for all matching points. The length of the line is proportional to the mismatch of the corresponding point. If the Error scaling factor is positive, the absolute error is displayed, if it is negative, the relative error is displayed. In the first case, the line length is equal to the Error scaling factor for the average error. In the second case, the line length is equal to the absolute value of the Error scaling factor for 100% error. To turn the error representation off, set the Error scaling factor equal to zero.
The standard weights used in MMP are defined in such a way that the absolute mismatching error along the boundaries is minimized in the least-squares sense. Therefore, you may obtain high relative errors in those areas where the field is relatively small. In order to reduce the high relative errors, you may increase the matching point weights in those areas. For doing this, you can define a function along the boundaries that specifies matching point dependent weights. For example, you may obtain the field along the boundaries from a first MMP computation without special weighting, when the MMP errors are computed and saved on a function file (Press the Save errors… button for saving.). Afterwards, you read the data in the function file into the function array ((Press the Read… button in the Function dialog.). Note that the array should have as many values as 2D matching points along the boundaries (see 2D box). In the Weight function arg. box you then define the number of the function array argument that shall be used for the special weighting. When you have saved the 2D errors of a previous MMP run, the first argument corresponds to the absolute errors, the second argument to the function values and the third argument to the relative errors. Therefore, you may set Weight function arg. 2 for a special weighting with the function values of the first run. You may also specify negative arguments in this box for inverting the weights.
Press the Points !! button, when you want to inspect the matching points generated by OpenMaXwell along the boundaries without setting up and solving the MMP matrix.
When you press the Solve !! button, OpenMaXwell will 1) generate the matching points, 2) set up and scale the rectangular MMP matrix (except when GUT is selected), 3) update and solve the triangular MMP matrix when Givens updating is selected or perform the CG iterations when CG is applied, 4) evaluate the amplitude if necessary, scale the MMP parameters, and compute the residual.
Press the Errors !! button when you want OpenMaXwell to evaluate the mismatching along the boundaries obtained from the current MMP expansions and MMP parameters defined in the Expansion dialog. This error analysis gives you a better impression of the accuracy of the results obtained from an MMP solution.
Pressing the Sol+err !! button is equivalent to pressing Solve !! followed by Errors!!
Press the Write err… (or Write errors…) button to write MMP error data to a function file. A file name dialog will allow you to select any file name. Note that the file will contain 1) the errors, 2) the field values, and 3) the relative errors in all matching points. The error and field components are compressed into a single real number.
You may save the 2D errors also for 3D models. For 3D models, each 3D matching point is associated with a 2D matching point that is used for generating the 3D points. The errors and field values in all 3D matching points correlated with a certain 2D matching point are averaged and associated to the 2D matching point. Saving the 2D errors for 3D models is important for adaptive expansion generation and for special matching point weighting.
Press the Read… button to read MMP data from a file. A file name dialog will allow you to select a file name. An MMP data file can contain rectangular or triangular MMP matrices. OpenMaXwell will ask whether you want to skip the corresponding data.
Press the Write… button to write MMP data to a file. A file name dialog will allow you to select a file name. When the rectangular or triangular MMP matrices have been computed, you can save them together with the MMP data defined in the dialog to the MMP file. OpenMaXwell will ask whether you want to save the matrix data.
To represent the rectangular or triangular MMP matrices graphically, you can store the MMP matrices in a *.FLD file. Press the Write Mtrx… button to write MMP matrices to a *.FLD file. A file name dialog will allow you to select a file name.
Press the Close button to close the dialog. Note that the current data in the dialog becomes effective when the dialog is closed.
Responsible for this web page: Ch. Hafner, Computational Optics Group, IEF, ETH, 8092 Zurich, Switzerland
Last update
31.03.2015