Static Phantom with off-resonance#
In this notebook, a simulation of a balanced steady state free precession with a spherical static phantom with air inclusions is performed. To do so the following steps are performed:
Define the Sequence (with cmrseq)
Define a 3D spherical phantom
Run simulation
Reconstruct images
Import modules and set TF-GPU configuration#
[1]:
from copy import deepcopy
import base64
from IPython.display import display, HTML, clear_output
# Load TF and check for GPUs
import tensorflow as tf
physical_devices = tf.config.list_physical_devices('GPU')
if physical_devices:
print("Available GPUS: \n\t", "\n\t".join([str(_) for _ in physical_devices]))
tf.config.experimental.set_visible_devices(physical_devices[0], 'GPU')
tf.config.experimental.set_memory_growth(physical_devices[0], True)
# 3rd Party dependencies
import cmrseq
from pint import Quantity
import pyvista
import numpy as np
from tqdm.notebook import tqdm
import matplotlib.pyplot as plt
%matplotlib inline
# Project library cmrsim
import sys
sys.path.insert(0, "../../")
import cmrsim
import cmrsim.utils.particle_properties as part_factory
Available GPUS:
PhysicalDevice(name='/physical_device:GPU:0', device_type='GPU')
2022-12-13 15:41:51.232566: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2022-12-13 15:41:52.188955: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1532] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 21480 MB memory: -> device: 0, name: NVIDIA TITAN RTX, pci bus id: 0000:b2:00.0, compute capability: 7.5
1. Define Sequence#
Configure Simulation Parameters#
[2]:
# Define MR-system specifications
system_specs = cmrseq.SystemSpec(max_grad=Quantity(50, "mT/m"), max_slew=Quantity(200., "mT/m/ms"),
grad_raster_time=Quantity(0.01, "ms"),
rf_raster_time=Quantity(0.01, "ms"),
adc_raster_time=Quantity(0.01, "ms"),
b0=Quantity(1.5, "T")
)
# Define Fourier-Encoding parameters
fov = Quantity([21, 21], "cm")
matrix_size = (151, 151)
res = fov / matrix_size
print("Resolution:", res)
n_dummy = 51
center_index = np.array(matrix_size) // 2 + np.mod(matrix_size, 2)
# Define imaging slice parameters
slice_thickness = Quantity(5, "mm")
slice_position = Quantity([0., 0., 0.], "m")
adc_duration=Quantity(2., "ms")
readout_direction = np.array([0., 1., 0.])
slice_normal = np.array([1., 0., 0.])
slice_position_offset = Quantity(0., "cm")
# Define RF-exication parameters (coupled with seeding slice atm)
flip_angle=Quantity(np.pi/4, "rad")
pulse_duration = Quantity(0.6, "ms")
Resolution: [0.1390728476821192 0.1390728476821192] centimeter
Instantiate Sequence#
Note: As a \(\alpha/2-TR/2\) preparation module is used, the first acquisition happens in the n_dummy+1 entry of the sequence list
[3]:
seq_list = cmrseq.seqdefs.sequences.balanced_ssfp(system_specs,
matrix_size,
inplane_resolution=res,
slice_thickness=slice_thickness,
adc_duration=adc_duration,
flip_angle=flip_angle,
pulse_duration=pulse_duration,
slice_position_offset=slice_position_offset,
dummy_shots=n_dummy)
/scratch/jweine/conda/envs/tf29_pyvista/lib/python3.9/site-packages/pint/quantity.py:1313: RuntimeWarning: invalid value encountered in double_scalars
magnitude = magnitude_op(new_self._magnitude, other._magnitude)
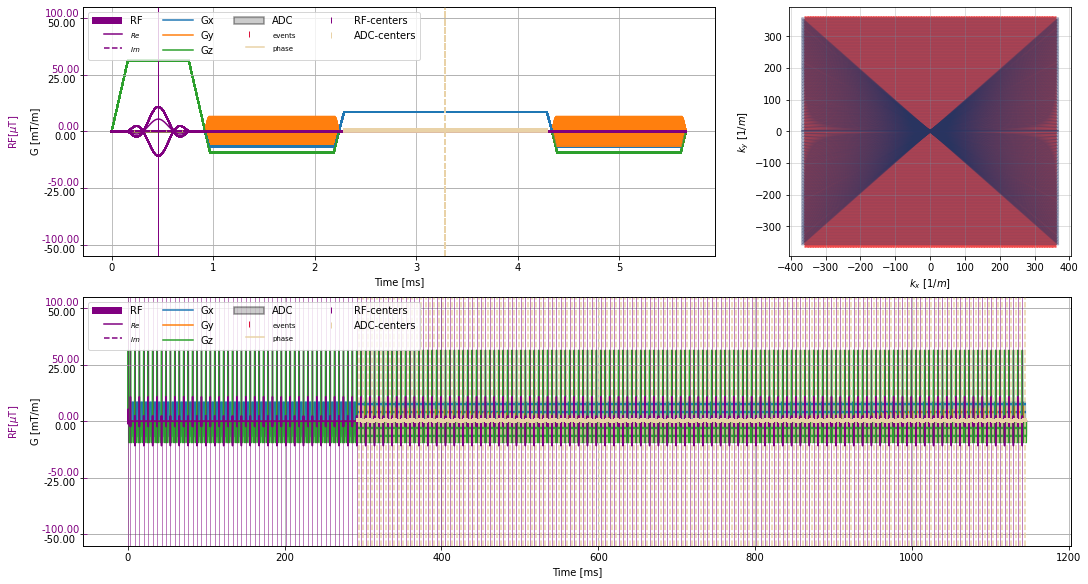
Plot sequence#
[4]:
_, k_adc, t_adc = seq_list[n_dummy + center_index[1]].calculate_kspace()
full_seq = deepcopy(seq_list[0])
full_seq.extend(deepcopy(seq_list[1:]))
clear_output()
plt.close("all")
f = plt.figure(constrained_layout=True, figsize=(15, 8))
axes = f.subplot_mosaic("AAB;CCC")
cmrseq.plotting.plot_sequence(seq_list[0], axes=axes["A"], format_axes=True, add_legend=False)
for s in seq_list[n_dummy:]:
cmrseq.plotting.plot_sequence(s, axes=[f.axes[-1], axes["A"], axes["A"], axes["A"]], format_axes=False)
for s in seq_list[n_dummy:]:
cmrseq.plotting.plot_kspace_2d(s, ax=axes["B"], k_axes=[0, 1])
cmrseq.plotting.plot_sequence(full_seq, axes=axes["C"])
2. Define Phantom#
Create 3D object#
[5]:
import sys
sys.path.append("../")
import local_functions
phantom = local_functions.create_spherical_phantom(dimensions=(61, 61, 61), spacing=(0.004, 0.004, 0.004))
clear_output()
# phantom = pyvista.UniformGrid(dimensions=(10, 20, 30), spacing=(0.004, 0.004, 0.004), origin=(-0.02, -0.04, -0.06))
# Create field to signal if regular mesh-points are within original unstructured grid
phantom["in_mesh"] = np.ones(phantom.points.shape[0])
# dataset = cmrsim.datasets.RegularGridDataset.from_shepp_logan(map_size=(100, 100, 50), extent=Quantity([20, 20, 10], "cm"))
# dataset.mesh["in_mesh"] = dataset.mesh["rho"] > 0
# dataset.mesh["M0"] = dataset.mesh["rho"]
Instantiate a RegularGridDataset#
Also assign MRI relevant material properties and comput the off-resonance distribution
[6]:
# Create regular grids
dataset = cmrsim.datasets.RegularGridDataset.from_unstructured_grid(phantom, pixel_spacing=Quantity([0.5, 0.5, 0.5], "mm"), padding=Quantity([5, 5, 5], "cm"))
# Assign properties (everything not 'in-mesh' is assumed to be air)
dataset.mesh["chi"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]) * (-9.05), np.ones_like(dataset.mesh["in_mesh"]) * 0.36) * 1e-6 # susceptibilty in ppm
dataset.mesh["M0"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]), np.zeros_like(dataset.mesh["in_mesh"])) # density in percent
dataset.mesh["T1"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]) * 1000, np.zeros_like(dataset.mesh["in_mesh"])) # time in ms
dataset.mesh["T2"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]) * 100, np.zeros_like(dataset.mesh["in_mesh"])) # time in ms
# Calculate b0-offresonance field
dataset.compute_offresonance(b0=system_specs.b0, susceptibility_key="chi");
/scratch/jweine/cmrsim/notebooks/bloch_simulation/../../cmrsim/datasets/_regular_grid.py:212: RuntimeWarning: invalid value encountered in true_divide
kernel = 1/3 - kz2 / (kx2 + ky2 + kz2)
/scratch/jweine/conda/envs/tf29_pyvista/lib/python3.9/site-packages/pyvista/core/dataset.py:1969: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
scalars = np.array(scalars)
Select a slice#
The slice coordinates are transformed into MPS coordinates as visualized below. The selected slice-mesh is wrapped again into a RegularGridDataset to subsequently extract the non-trivial material points as dictionary of arrays.
[7]:
slice_mesh = dataset.select_slice(slice_normal=slice_normal, spacing=Quantity((0.25, 0.25, 1.5), "mm"),
slice_position=slice_normal*slice_position_offset,
field_of_view=Quantity([22, 22, slice_thickness.m_as("cm")*1.5], "cm"),
readout_direction=readout_direction, in_mps=True)
slice_dataset = cmrsim.datasets.RegularGridDataset(slice_mesh)
phantom_dict = slice_dataset.get_phantom_def(filter_by="in_mesh", keys=["M0", "T1", "T2", "offres"],
perturb_positions_std=Quantity(0.005, "mm").m_as("m"))
/scratch/jweine/cmrsim/notebooks/bloch_simulation/../../cmrsim/datasets/_regular_grid.py:122: UserWarning: Optimal rotation is not uniquely or poorly defined for the given sets of vectors.
rot, _ = Rotation.align_vectors(original_basis, new_basis)
Plot 3D phantom with offresonance map
[8]:
import imageio
pyvista.close_all()
pyvista.start_xvfb()
plotter = pyvista.Plotter(notebook=False, off_screen=True, window_size=(600, 400))
plotter.add_mesh(dataset.mesh.threshold(0.1, "M0"), scalars="offres", opacity=0.65,
cmap="twilight", scalar_bar_args=dict(title="Offresonance (T)"))
local_functions.add_custom_axes(plotter, scale=0.3)
box = local_functions.transformed_box(np.eye(3, 3), slice_normal, readout_direction, slice_position_offset*slice_normal,
extend=Quantity([*fov.m_as("m"), slice_thickness.m_as("m")], "m"))
plotter.add_mesh(box)
plotter.screenshot("polydata_temp0.png")
plotter.close()
plotter = pyvista.Plotter(notebook=False, off_screen=True, window_size=(600, 400))
plotter.add_mesh(pyvista.PolyData(phantom_dict["r_vectors"]), scalars=phantom_dict["offres"], opacity=0.65, cmap="twilight", scalar_bar_args=dict(title="Offresonance (T)"))
local_functions.add_custom_axes(plotter, scale=0.3)
plotter.screenshot("polydata_temp1.png")
img = np.concatenate([imageio.v3.imread("polydata_temp0.png"), imageio.v3.imread("polydata_temp1.png")], axis=1)
imageio.v3.imwrite("polydata_temp.png", img)
b64 = base64.b64encode(open("polydata_temp.png",'rb').read()).decode('ascii')
display(HTML(f'<img src="data:image/gif;base64,{b64}" />'))
/scratch/jweine/cmrsim/notebooks/bloch_simulation/../local_functions.py:41: UserWarning: Optimal rotation is not uniquely or poorly defined for the given sets of vectors.
rot, _ = Rotation.align_vectors(reference_basis, new_basis)
3. Perfom Simulation#
Instantiate simulation modules#
From the preparation phase as well as the acquisition TRs grid the sequence and instantiate the modules including the off-resonance submodule
[9]:
dummy_sequence = deepcopy(seq_list[0])
dummy_sequence.extend(seq_list[1:n_dummy+1])
time_dummy, rf_grid_dummy, grad_grid_dummy, _ = [np.stack(v) for v in cmrseq.utils.grid_sequence_list([dummy_sequence, ])]
time, rf_grid, grad_grid, adc_on_grid = [np.stack(v) for v in cmrseq.utils.grid_sequence_list(seq_list[n_dummy+1:])]
print(time_dummy.shape, rf_grid_dummy.shape, grad_grid_dummy.shape)
print(time.shape, rf_grid.shape, grad_grid.shape, adc_on_grid.shape)
# Define off-resonsance submodule
offres = cmrsim.bloch.submodules.OffResonance(gamma=system_specs.gamma.m_as("MHz/T"), device="GPU:0")
# Construct BlochOperators and pass offresonance module:
module_dummyshots = cmrsim.bloch.GeneralBlochOperator(name="dummy_shots", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
time_grid=time_dummy[0],
gradient_waveforms=grad_grid_dummy,
rf_waveforms=rf_grid_dummy,
device="GPU:0",
submodules=[offres]
)
module_acquisition = cmrsim.bloch.GeneralBlochOperator(name="acquisition", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
time_grid=time[0],
gradient_waveforms=grad_grid,
rf_waveforms=rf_grid,
adc_events=adc_on_grid,
device="GPU:0",
submodules=[offres]
)
Extending Sequence: 100%|████████████████████████████████████████████████████████████████████████████████████████████████████| 51/51 [00:00<00:00, 322.52it/s]
(1, 29150) (1, 29150) (1, 29150, 3)
(151, 717) (151, 717) (151, 717, 3) (151, 717, 2)
Create Bloch-dataset for batched data stream#
[10]:
properties = {"M0": phantom_dict["M0"].astype(np.float32),
"T1": phantom_dict["T1"].astype(np.float32),
"T2": phantom_dict["T2"].astype(np.float32),
"off_res": (phantom_dict["offres"]).astype(np.float32).reshape(-1, 1),
"magnetization": part_factory.norm_magnetization()(phantom_dict["M0"].shape[0]),
"initial_position": phantom_dict["r_vectors"].astype(np.float32)
}
input_dataset = cmrsim.datasets.BlochDataset(properties, filter_inputs=True)
print(properties["M0"].shape)
(2414860,)
Call modules in loop-structure#
[11]:
# Instantiate simulator:
print(f"Total time-steps per TR: {time.shape[1]}")
for batch in tqdm(input_dataset(batchsize=int(3e6)).take(1)):
print({k:v.shape for k, v in batch.items()})
m_init = batch.pop("magnetization")
initial_position = batch.pop("initial_position")
m, r = module_dummyshots(initial_position=initial_position, magnetization=m_init, **batch)
# m, r = m_init, initial_position
for tr_index in tqdm(range(time.shape[0]), desc="Iterating TRs", leave=False):
m, r = module_acquisition(initial_position=r, magnetization=m, repetition_index=tr_index, **batch)
Total time-steps per TR: 717
{'M0': TensorShape([2414860]), 'T1': TensorShape([2414860]), 'T2': TensorShape([2414860]), 'off_res': TensorShape([2414860, 1]), 'magnetization': TensorShape([2414860, 3]), 'initial_position': TensorShape([2414860, 3])}
2022-12-13 15:44:21.659296: I tensorflow/compiler/xla/service/service.cc:170] XLA service 0x7fcd9c02c280 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices:
2022-12-13 15:44:21.659362: I tensorflow/compiler/xla/service/service.cc:178] StreamExecutor device (0): NVIDIA TITAN RTX, Compute Capability 7.5
2022-12-13 15:44:21.669559: I tensorflow/compiler/mlir/tensorflow/utils/dump_mlir_util.cc:263] disabling MLIR crash reproducer, set env var `MLIR_CRASH_REPRODUCER_DIRECTORY` to enable.
2022-12-13 15:44:21.772288: I tensorflow/core/platform/default/subprocess.cc:304] Start cannot spawn child process: No such file or directory
2022-12-13 15:44:22.383555: I tensorflow/compiler/jit/xla_compilation_cache.cc:478] Compiled cluster using XLA! This line is logged at most once for the lifetime of the process.
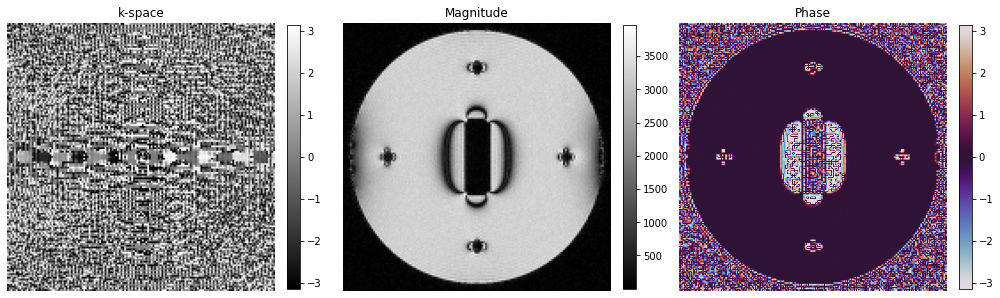
4. Reconstruct images#
[12]:
time_signal = tf.stack(module_acquisition.time_signal_acc, axis=0).numpy().reshape(matrix_size[::-1], order="F")
time_signal += tf.complex(*[tf.random.normal(shape=time_signal.shape, stddev=50) for i in range(2)])
centered_projection = tf.signal.fft(time_signal).numpy()
centered_k_space = tf.signal.fft(centered_projection).numpy()
image = tf.signal.fftshift(tf.signal.ifft2d(tf.roll(tf.signal.ifftshift(centered_k_space, axes=(0, 1)), -1, axis=1)), axes=(0, 1)).numpy()
[13]:
# Show images
fig, axes = plt.subplots(1, 3, figsize=(14, 5))
# kspace_plot = axes[0].imshow(np.log(np.abs(np.squeeze(centered_k_space))), cmap="gray")
kspace_plot = axes[0].imshow(np.angle(np.squeeze(centered_k_space)), cmap="gray")
fig.colorbar(kspace_plot, ax=axes[0], fraction=0.045, pad=0.04)
abs_plot = axes[1].imshow(np.abs(np.squeeze(image)), cmap="gray")
fig.colorbar(abs_plot, ax=axes[1], fraction=0.045, pad=0.04)
phase_plot = axes[2].imshow(np.angle(np.squeeze(image)), cmap="twilight", vmin=-np.pi, vmax=np.pi)
fig.colorbar(phase_plot, ax=axes[2], fraction=0.045, pad=0.04)
[_.axis("off") for _ in axes]
[_.set_title(t) for _, t in zip(axes, ["k-space", "Magnitude", "Phase"])]
fig.tight_layout()