LV Contraction and breathing motion with bSSFP#
This notebook contains the code to reproduce the introductory example described in the CMRseq publication.
At the end of this notebook, the non-executable code excerpt included in the paper is appended.
Imports#
[1]:
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "1"
import tensorflow as tf
print(tf.config.get_visible_devices("GPU"))
gpu = tf.config.get_visible_devices("GPU")[1]
tf.config.set_visible_devices(gpu, device_type="GPU")
tf.config.experimental.set_memory_growth(gpu, True)
from IPython.display import display, Image, HTML, clear_output
import sys
import math
import itertools
import base64
from typing import List, Union, Tuple, Sequence
from pint import Quantity
from scipy.optimize import curve_fit
import pyvista
import vtk
from tqdm.notebook import tqdm
import ipywidgets
import numpy as np
import matplotlib.pyplot as plt
sys.path.insert(0, "../../../cmrseq/")
sys.path.insert(0, "../../../cmrsim/")
import cmrsim
import cmrseq
2023-08-31 10:20:08.851313: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
[PhysicalDevice(name='/physical_device:GPU:0', device_type='GPU'), PhysicalDevice(name='/physical_device:GPU:1', device_type='GPU')]
Load Phantom#
All phantom snapshots are saved as vtk file. The CardiacMeshDataset is used to load all provided files and store the snapshots as displacements in a single unstructured grid.
[2]:
shared_drive_path = "/mnt/itetstor/jweine/mritrans_ibtscratch/"
mesh_resource = f"{shared_drive_path}/Stefano/ForJonathan"
[ ]:
files = [f'{mesh_resource}/NOR_new_1_ED_reference/Displacements/Displ{i:03}.vtk' for i in range(0, 228, 1)]
timing = Quantity(np.loadtxt(f"{mesh_resource}/NOR_new_1_ED_reference/PV_loop_reordered.txt")[:, 1], "ms")
refine_module = cmrsim.datasets.CardiacMeshDataset.from_list_of_meshes(files, timing[:len(files)], time_precision_ms=3, mesh_numbers=(4, 40, 30))
Refine mesh#
To reduce discretization artifacts, the mesh is refined at each snapshot. As intermediate result, the single vtk is saved.
[ ]:
refined_mesh = refine_module.refine(longitudinal_factor=5, circumferential_factor=4, radial_factor=5)
refined_mesh.mesh.save(f"refined_mesh_{tuple(refined_mesh.mesh_numbers[i] for i in (2, 0, 1))}.vtk")
Save cell volumes#
First the previously refined and saved mesh is loaded, and subsequently the cell sizes are converted to factors describing the available equlibrium magnetization per mesh node.
[3]:
fn = "refined_mesh_(16, 161, 150).vtk"
mesh_numbers = [int(s) for s in fn.replace("refined_mesh_(", "").replace(").vtk", "").split(",")]
refined_mesh = cmrsim.datasets.CardiacMeshDataset.from_single_vtk(f"{fn}", time_precision_ms=3, mesh_numbers=mesh_numbers)
[ ]:
ref_time, ref_pos = refined_mesh.get_trajectories(start=Quantity(0, "ms"), end=Quantity(refined_mesh.timing[-1] + 1, "ms"))
connectivity = refined_mesh.mesh.cells_dict[10].reshape(-1, 4)
proton_density_weights = np.stack([cmrsim.datasets.CardiacMeshDataset.evaluate_cellsize(ref_pos[:, i].m_as("m"), connectivity)[2]
for i in tqdm(range(ref_time.shape[0]), desc="Calculating cell volumes", leave=False)])[..., np.newaxis]
np.save("pd.npy", proton_density_weights)
Inititalize Trajectory modules#
The following three modules are instantiated: 1. M0 - interpolation 2. Particle position due to contraction 3. Breathing wrapper
[4]:
ref_time, ref_pos = refined_mesh.get_trajectories(start=Quantity(0, "ms"), end=Quantity(refined_mesh.timing[-1] + 1, "ms"))
proton_density_weights = np.load("pd.npy")
batch_size=400_000
proton_density_module = cmrsim.trajectory.PODTrajectoryModule(ref_time.m_as("ms"), proton_density_weights.transpose(1, 0, 2),
n_modes=10, poly_order=10, batch_size=batch_size, is_periodic=True)
contraction_module = cmrsim.trajectory.PODTrajectoryModule(ref_time.m_as("ms"), ref_pos.m_as("m"),
n_modes=10, poly_order=10,
batch_size=batch_size, is_periodic=True)
breathing_module = cmrsim.trajectory.SimpleBreathingMotionModule.from_sinosoidal_motion(sub_trajectory=contraction_module,
breathing_period=Quantity(7, "s"),
breathing_direction=np.array([1, 1, 3]),
breathing_amplitude=Quantity(2.5, "cm"))
proton_density_weights.shape, proton_density_weights.dtype, ref_time.shape, ref_time.dtype
/opt/conda/lib/python3.11/site-packages/numpy/polynomial/polyutils.py:660: RuntimeWarning: overflow encountered in square
scl = np.sqrt(np.square(lhs).sum(1))
/opt/conda/lib/python3.11/site-packages/numpy/core/_methods.py:49: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/opt/conda/lib/python3.11/site-packages/numpy/polynomial/polynomial.py:1362: RankWarning: The fit may be poorly conditioned
return pu._fit(polyvander, x, y, deg, rcond, full, w)
[4]:
((226, 386416, 1), dtype('float64'), (226,), dtype('float64'))
Define Sequence#
The cmrseq package is used for sequence definition.
[5]:
# Define MR-system specifications
system_specs = cmrseq.SystemSpec(max_grad=Quantity(50, "mT/m"), max_slew=Quantity(200., "mT/m/ms"),
grad_raster_time=Quantity(0.01, "ms"),
rf_raster_time=Quantity(0.01, "ms"),
adc_raster_time=Quantity(0.01, "ms"),
b0=Quantity(1.5, "T")
)
# Define Fourier-Encoding parameters
fov = Quantity([15, 15], "cm")
matrix_size = (101, 101)
n_dummy=81
seq_list = cmrseq.seqdefs.sequences.balanced_ssfp(system_specs,
matrix_size,
repetition_time=Quantity(0., "ms"),
inplane_resolution=fov / matrix_size,
slice_thickness=Quantity(5, "mm"),
adc_duration=Quantity(2., "ms"),
flip_angle=Quantity(np.pi/4, "rad"),
pulse_duration=Quantity(0.6, "ms"),
slice_position_offset=Quantity(0., "cm"),
dummy_shots=n_dummy)
/scratch/jweine/cmrsim/notebooks/bloch_simulation/../../../cmrseq/cmrseq/parametric_definitions/sequences/_ssfp.py:129: UserWarning: Repetition time too short to be feasible, set TR to 4.16 millisecond
warn(f"Repetition time too short to be feasible, set TR to {minimal_tr}")
[58]:
_, k_adc, t_adc = seq_list[n_dummy + 51].calculate_kspace()
full_seq = seq_list[0].copy()
full_seq.extend(seq_list[1:])
plt.close("all")
f = plt.figure(constrained_layout=True, figsize=(15, 8))
axes = f.subplot_mosaic("AAB;CCC")
cmrseq.plotting.plot_sequence(seq_list[0], axes=axes["A"], format_axes=True, add_legend=False)
for s in tqdm(seq_list[n_dummy:]):
cmrseq.plotting.plot_sequence(s, axes=[f.axes[-1], axes["A"], axes["A"], axes["A"]], format_axes=False)
for s in tqdm(seq_list[n_dummy:]):
cmrseq.plotting.plot_kspace_2d(s, ax=axes["B"], k_axes=[0, 1])
cmrseq.plotting.plot_sequence(full_seq, axes=axes["C"])
f.savefig("sequence_plot.png", dpi=400)
clear_output()
display(Image("sequence_plot.png"))
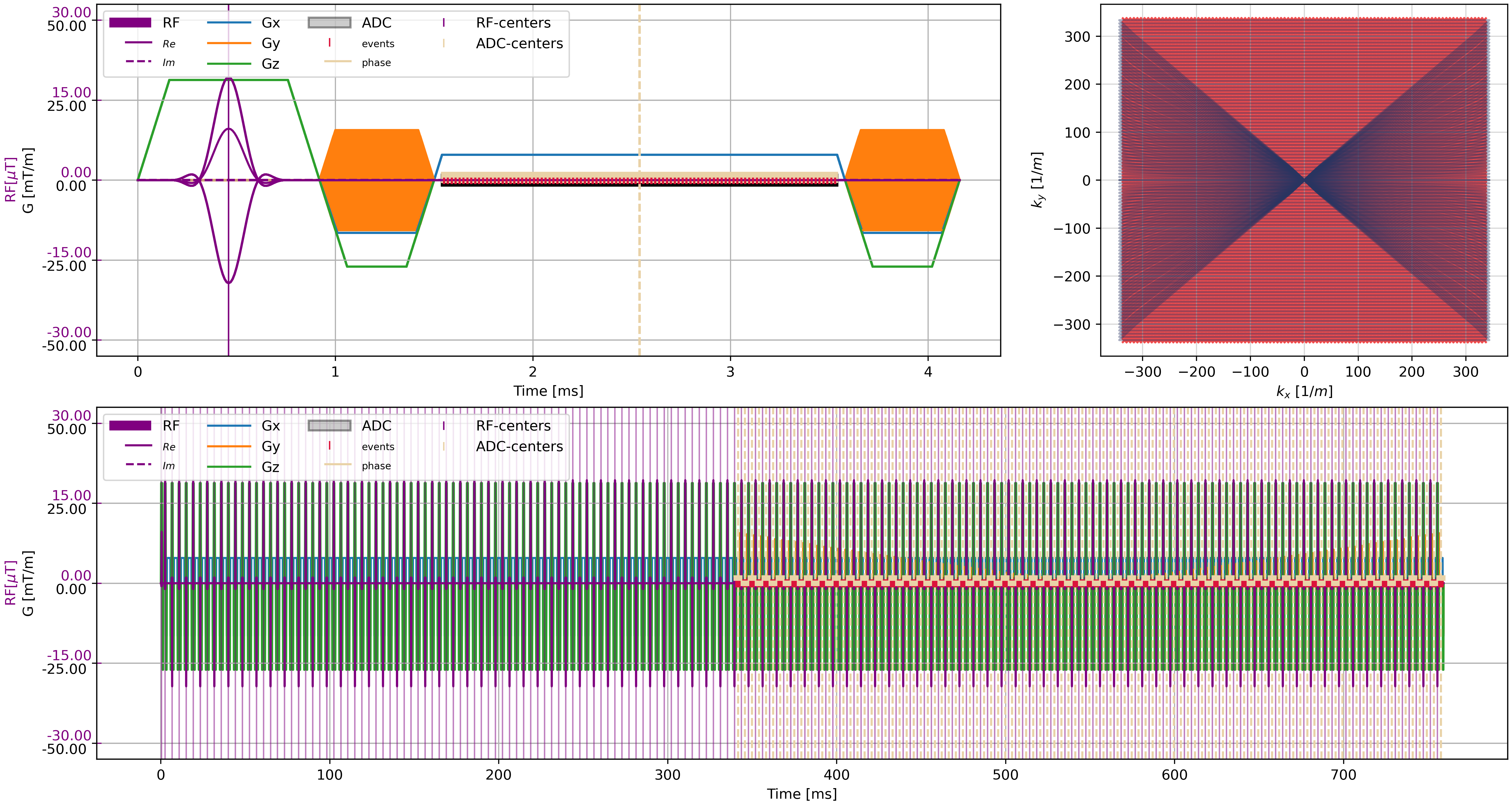
Simulation#
Initialize Bloch operators#
Again cmrseq is used to create the gridded version of all waveforms.
[6]:
dummy_sequence = seq_list[0].copy()
dummy_sequence.extend(seq_list[1:n_dummy+1])
time_dummy, rf_grid_dummy, grad_grid_dummy, _ = [np.stack(v) for v in cmrseq.utils.grid_sequence_list([dummy_sequence, ])]
time, rf_grid, grad_grid, adc_on_grid = [np.stack(v) for v in cmrseq.utils.grid_sequence_list(tqdm(seq_list[n_dummy+1:]))]
print(time_dummy.shape, rf_grid_dummy.shape, grad_grid_dummy.shape)
print(time.shape, rf_grid.shape, grad_grid.shape, adc_on_grid.shape)
# Construct BlochOperators and pass offresonance module:
module_dummyshots = cmrsim.bloch.GeneralBlochOperator(name="dummy_shots", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
time_grid=time_dummy[0],
gradient_waveforms=grad_grid_dummy,
rf_waveforms=rf_grid_dummy,
device="GPU:0",
)
module_acquisition = cmrsim.bloch.GeneralBlochOperator(name="acquisition", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
time_grid=time[0],
gradient_waveforms=grad_grid,
rf_waveforms=rf_grid,
adc_events=adc_on_grid,
device="GPU:0",
)
Extending Sequence: 100%|██████████| 81/81 [00:00<00:00, 263.49it/s]
(1, 33905) (1, 33905) (1, 33905, 3)
(101, 517) (101, 517) (101, 517, 3) (101, 517, 2)
Setup input data#
[7]:
end_dummy = time_dummy[0, -1]
start_of_trs = (np.array([-end_dummy, 0, *np.cumsum(time[:, -1]).tolist()]) + end_dummy).astype(np.float32)
M0_initial = np.concatenate([proton_density_module(None, start_of_trs[0], batch_index=b)[0][:, 0, 0] for b in range(3)], axis=0)
properties = {"M0": M0_initial,
"T1": np.ones_like(M0_initial) * 1000,
"T2": np.ones_like(M0_initial) * 200,
"magnetization": cmrsim.utils.particle_properties.norm_magnetization()(M0_initial.shape[0]),
"initial_position": ref_pos[:, 0].m_as("m").astype(np.float32)
}
input_dataset = cmrsim.datasets.BlochDataset(properties, filter_inputs=False)
print(properties["M0"].shape, properties["initial_position"].shape)
(386416,) (386416, 3)
Run#
To change the case of motion, pass the corresponding trajectory module to the bloch-module call-invokes.
[9]:
tf.config.run_functions_eagerly(False)
print(f"Total time-steps per TR: {time.shape[1]}")
module_acquisition.reset()
for batch_index, batch in tqdm(input_dataset(batchsize=int(contraction_module.batch_size.read_value())).enumerate()):
breathing_module.current_time_ms.assign(0.)
contraction_module._taylor_module.current_time_ms.assign(0.)
m_init = batch.pop("magnetization")
initial_position = batch.pop("initial_position")
m0 = proton_density_module(initial_position, start_of_trs[0], batch_index=tf.cast(batch_index, tf.int32))[0][:, 0, 0]
batch["M0"] = m0
print({k:v.shape for k, v in batch.items()}, {"r":initial_position.shape, "m":m_init.shape})
contraction_module.current_batch_idx.assign(tf.cast(batch_index, dtype=tf.int32))
m, r = module_dummyshots(initial_position=initial_position, magnetization=m_init, **batch,
trajectory_module=contraction_module,
# trajectory_module=breathing_module
)
pbar = tqdm(range(time.shape[0]), desc="Iterating TRs", leave=False)
for tr_index in pbar:
pbar.set_postfix_str(f"Current simulation time: {contraction_module.current_time_ms.read_value(): 1.3f}")
m0 = proton_density_module(initial_position, start_of_trs[tr_index], batch_index=tf.cast(batch_index, tf.int32))[0][:, 0, 0]
batch["M0"] = m0
m, r = module_acquisition(initial_position=r, magnetization=m, repetition_index=tr_index, **batch,
trajectory_module=contraction_module,
# trajectory_module=breathing_module
)
Total time-steps per TR: 517
{'M0': TensorShape([386416]), 'T1': TensorShape([386416]), 'T2': TensorShape([386416])} {'r': TensorShape([386416, 3]), 'm': TensorShape([386416, 3])}
Reconstruction#
[10]:
time_signal = tf.stack(module_acquisition.time_signal_acc, axis=0).numpy().reshape(matrix_size[::-1], order="F")
time_signal += tf.complex(*[tf.random.normal(shape=time_signal.shape, stddev=30) for i in range(2)])
centered_projection = tf.signal.fft(time_signal).numpy()
centered_k_space = tf.signal.fft(centered_projection).numpy()
image = tf.signal.fftshift(tf.signal.ifft2d(tf.roll(tf.signal.ifftshift(centered_k_space, axes=(0, 1)), -1, axis=1)), axes=(0, 1)).numpy()
Depending on which motion case is coosen, save the intermediate result
[11]:
# np.savez("no_motion_result.npz", time_signal=time_signal.numpy(), image=image)
np.savez("contraction_motion_result.npz", time_signal=time_signal.numpy(), image=image)
# np.savez("breathing_motion_result.npz", time_signal=time_signal.numpy(), image=image)
Create figure for publication#
[ ]:
start, end = 0., contraction_module._taylor_module.current_time_ms.read_value().numpy()
pos, _ = breathing_module(ref_pos.m, np.array([0., 750.], dtype=np.float32))
temp_mesh_start = pyvista.UnstructuredGrid(refined_mesh.mesh.cells_dict, pos[:, 0].numpy())
temp_mesh_end = pyvista.UnstructuredGrid(refined_mesh.mesh.cells_dict, pos[:, 1].numpy())
coms = np.mean(pos, axis=0)
sphere1 = pyvista.Sphere(radius=0.001, center=coms[0])
sphere2 = pyvista.Sphere(radius=0.001, center=coms[1])
pyvista.close_all()
pyvista.start_xvfb()
pyvista.set_jupyter_backend("panel")
# plotter = pyvista.Plotter(off_screen=False, window_size=(800, 800))
plotter = pyvista.Plotter(off_screen=True, window_size=(800, 800))
plotter.add_mesh(temp_mesh_start, show_scalar_bar=False, color="lightblue", opacity=0.2)
plotter.add_mesh(sphere1, color="lightblue")
plotter.add_mesh(temp_mesh_end, show_scalar_bar=False, color="lightgreen", opacity=0.2)
plotter.add_mesh(sphere2, color="lightgreen")
plotter.add_arrows(coms[0:1], coms[1:2] - coms[:1], color="red")
plotter.camera_position = "xz"
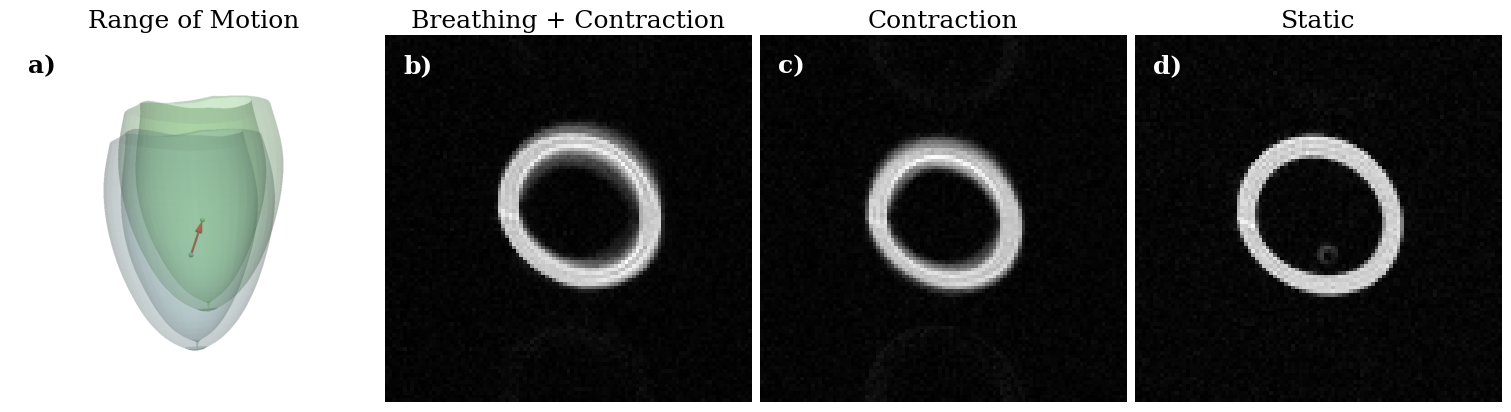
plotter.screenshot("range_of_motion.png", window_size=(1000, 1000));
plotter.close()
[13]:
import imageio
plt.rcParams["font.family"] = "serif"
figure, axes = plt.subplots(1, 4, constrained_layout=True, figsize=(15, 5))
[ax.set_title(t, fontsize=18) for ax, t in zip(axes, ["Range of Motion", "Breathing + Contraction", "Contraction", "Static"])]
[ax.axis("off") for ax in axes]
axes[0].imshow(imageio.v3.imread("range_of_motion.png"))
axes[1].imshow(np.abs(np.load("breathing_motion_result.npz")["image"]), cmap="gray")
axes[2].imshow(np.abs(np.load("contraction_motion_result.npz")["image"]), cmap="gray")
axes[3].imshow(np.abs(np.load("no_motion_result.npz")["image"]), cmap="gray")
[ax.text(0.05, 0.95, f"{t})", horizontalalignment='left', verticalalignment='top', transform = ax.transAxes,
fontsize=18, fontweight="bold", color=c) for ax, t, c in zip(axes, "abcd", "kwww")]
figure.savefig("Figure5.tiff", dpi=500)
# figure.savefig("movemesh_bssfp.png", dpi=300)
Paper non-executable code#
[ ]:
files = [f'{resource_path}/Displ{i:03}.vtk' for i in range(0, 228, 1)]
timing = Quantity(np.loadtxt(f"{resource_path}/snapshot_timings.txt"), "ms")
refine_module = cmrsim.datasets.CardiacMeshDataset.from_list_of_meshes(files, timing, ...)
refined_mesh = refine_module.refine(longitudinal_factor=5, circumferential_factor=4, radial_factor=5)
ref_time, ref_pos = refined_mesh.get_trajectories(...)
pd_weights_t0 = cmrsim.datasets.CardiacMeshDataset.evaluate_cellsize(ref_pos[:, 0].m_as("m"), connectivity)
...
proton_density_weights = np.stack([pd_weights_t0, ...])
[ ]:
batch_size=200_000
proton_density_module = cmrsim.trajectory.PODTrajectoryModule(ref_time.m_as("ms"), proton_density_weights,
n_modes=10, poly_order=10, batch_size=batch_size, is_periodic=True)
contraction_module = cmrsim.trajectory.PODTrajectoryModule(ref_time.m_as("ms"), ref_pos.m_as("m"),
n_modes=10, poly_order=10, batch_size=batch_size, is_periodic=True)
breathing_module = cmrsim.trajectory.SimpleBreathingMotionModule(contraction_module, breathing_curve, ...)
module_acquisition = cmrsim.bloch.GeneralBlochOperator(time_grid=time, gradient_waveforms=grad_grid,
rf_waveforms=rf_grid, adc_events=adc_on_grid, ...)
[ ]:
for batch_index, batch in tqdm(input_dataset):
m0 = proton_density_module(initial_position, start_of_trs[0], batch_index=batch_index)
batch["M0"] = m0
for tr_index in range(n_tr):
m0 = contraction_module(initial_position, start_of_trs[tr_index], batch_index=tf.cast(batch_index, tf.int32))[0][:, 0, 0]
batch["M0"] = m0
m, r = module_acquisition(initial_position=r, magnetization=m, repetition_index=tr_index,
trajectory_module=breathing_module, **batch)