Represent Trajectory with their Taylor Expansion#
In this notebook, the functionality of the TaylorTrajectory module is hightlighted using two examples. First a toy example of a particles with artificial kinematics is used. Secondly, a slice of a contracting LV is used as input.
Imports#
[2]:
import sys
sys.path.append("../")
sys.path.insert(0, "../../")
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
os.environ['CUDA_VISIBLE_DEVICES'] = "3"
import tensorflow as tf
gpu = tf.config.get_visible_devices("GPU")
if gpu:
tf.config.experimental.set_memory_growth(gpu[0], True)
[3]:
from IPython.display import display, HTML, clear_output
import base64
import imageio
import pyvista
import numpy as np
from tqdm.notebook import tqdm
import matplotlib.pyplot as plt
from pint import Quantity
import cmrsim
import local_functions
Simple example#
Construct trajectories#
[11]:
n_p = 50
n_t = 50
r0 = Quantity(np.random.normal(0., 0.001, size=(n_p, 3)), "m")
t = Quantity(np.linspace(0, 100, n_t), "ms")
v = Quantity(np.random.uniform(1, 2, size=(n_p, 3)), "cm/s")
a = Quantity(np.random.uniform(1, 2, size=(n_p, 3)), "cm/s**2")
r = a.reshape(n_p, 1, 3) * t.reshape(1, n_t, 1)**2 + v.reshape(n_p, 1, 3) * t.reshape(1, n_t, 1) + r0.reshape(n_p, 1, 3)
Fit trajectory Module#
[12]:
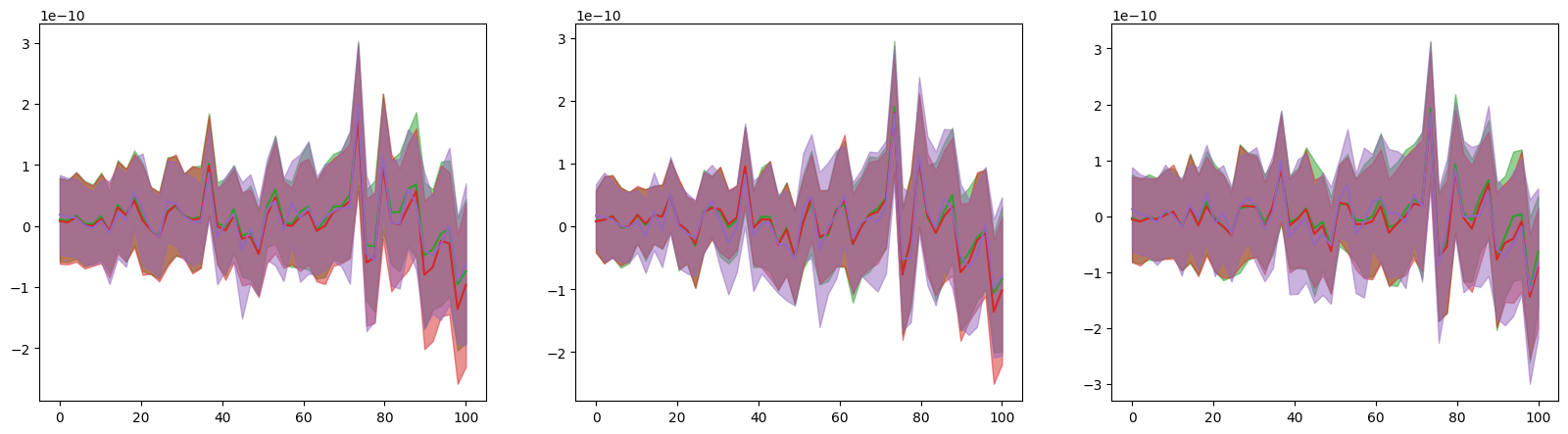
fig, ax = plt.subplots(1, 3, figsize=(20, 5))
for order in range(2, 5):
trajectory_module = cmrsim.trajectory.TaylorTrajectoryN(order=order, time_grid=t.m_as("ms"), particle_trajectories=r.m_as("m"))
taylor_positions, _ = trajectory_module(None, t.m_as("ms").astype(np.float32))
m1 = np.mean(taylor_positions - r.m_as("m"), axis=0)
s1 = np.std(taylor_positions - r.m_as("m"), axis=0)
for i in range(3):
ax[i].plot(t.m_as("ms"), m1[:, i], color=f"C{order}")
ax[i].fill_between(t.m_as("ms"), m1[:, i] - s1[:, i], m1[:, i] + s1[:, i], color=f"C{order}", alpha=0.5)
WARNING:tensorflow:5 out of the last 5 calls to <function TaylorTrajectoryN._evaluate_trajectory at 0x7f62c4600a40> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has reduce_retracing=True option that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
WARNING:tensorflow:6 out of the last 6 calls to <function TaylorTrajectoryN._evaluate_trajectory at 0x7f62c4600040> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has reduce_retracing=True option that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
Visualize results#
[13]:
# Animate trajectories
pyvista.start_xvfb()
plotter = pyvista.Plotter(off_screen=True, window_size=(600, 600))
local_functions.animate_trajectories(plotter, np.swapaxes(r.m_as("m"), 0, 1), t.m_as("ms"), filename="particles_test.gif",
scalars=np.swapaxes(taylor_positions-r.m_as("m"), 0, 1),
text_kwargs=dict(color="black", font_size=12),
mesh_kwargs=dict(show_scalar_bar=True, color="gray", render_points_as_spheres=True, cmap="viridis",
scalar_bar_args={"title": "Displacement to step [m]", "position_y": 0.05}),
visualize_trajectories=True,
trajectory_kwargs=dict(cmap="hot", scalar_bar_args={"title": "Displacement per step [m]", "position_y": 0.9}))
plotter.close()
# Show final state
pyvista.close_all()
t_idx = -1
plot_mesh = pyvista.PolyData(taylor_positions[:, t_idx].numpy())
ref_mesh = pyvista.PolyData(r[:, t_idx].m_as("m"))
plot_mesh["disp"] = ref_mesh.points - plot_mesh.points
glyphs = plot_mesh.glyph(orient="disp", geom=pyvista.Line())
plotter = pyvista.Plotter(off_screen=True, window_size=(600, 600))
plotter.add_mesh(plot_mesh, render_points_as_spheres=True, show_scalar_bar=False, color="white")
plotter.add_mesh(ref_mesh, render_points_as_spheres=True, show_scalar_bar=False, color="steelblue")
plotter.add_mesh(glyphs, cmap="hot", show_scalar_bar=True, scalar_bar_args=dict(title="Difference to ref (m)"))
plotter.add_title("Final Difference to ref")
img = plotter.screenshot()
# Combine figures and display
gif1 = imageio.v3.imread("particles_test.gif")
stacked_frames = [np.concatenate([frame, img], axis=1) for frame in gif1]
imageio.mimsave("particles_testc.gif", stacked_frames, loop=10, duration=0.3)
b64 = base64.b64encode(open("particles_testc.gif", 'rb').read()).decode('ascii')
display(HTML(f'<img src="data:image/gif;base64,{b64}" />'))
LV-mesh example#
Load determinisitic trajectories#
The node-trajectories of a sliced contracting left ventricle mesh are used to define the input for the trajectory module.
[17]:
!wget https://gitlab.ethz.ch/ibt-cmr/modeling/cmr-random-diffmaps/-/archive/main/cmr-random-diffmaps-main.zip?path=notebooks/example_data -O test_data.zip
!unzip test_data.zip
!rm test_data.zip -r
!mv cmr-random-diffmaps-main-notebooks-example_data/notebooks/example_data test_data
!rm cmr-random-diffmaps-main-notebooks-example_data -r
clear_output()
[18]:
files = [f'test_data/example_mesh/Displacements/Displ{i:03}.vtk' for i in range(0, 100, 1)]
timing = Quantity(np.loadtxt("test_data/example_mesh/PV_loop_reordered.txt")[:, 1], "ms")
original_mesh = cmrsim.datasets.CardiacMeshDataset.from_list_of_meshes(files, timing[:len(files)],
mesh_numbers=(4, 40, 30),
time_precision_ms=3)
refined_mesh = original_mesh.refine(longitudinal_factor=2, circumferential_factor=1, radial_factor=1)
slice_dict = dict(slice_thickness=Quantity(3, "mm"), slice_normal=np.array((0., 0., 1.)),
slice_position=Quantity([0, 0, -3], "cm"), reference_time=Quantity(105, "ms"))
temp = refined_mesh.select_slice(**slice_dict)
slice_mesh = cmrsim.datasets.MeshDataset(temp, refined_mesh.timing)
Loading Files... : 100%|██████████| 99/99 [00:01<00:00, 51.64it/s]
Refining mesh by factors: l=2, c=1, r=1: 100%|██████████| 100/100 [00:22<00:00, 4.51it/s]
[19]:
t, r = slice_mesh.get_trajectories(start=Quantity(105, "ms"), end=Quantity(300, "ms"))
np.min(t), np.max(t), r.shape, r.units
[19]:
(105.045 <Unit('millisecond')>,
298.975 <Unit('millisecond')>,
(624, 49, 3),
<Unit('meter')>)
Visualize the reference trajectories#
[20]:
pyvista.start_xvfb()
plotter = pyvista.Plotter(off_screen=True, window_size=(500, 500))
local_functions.animate_trajectories(plotter, np.swapaxes(r.m_as("m"), 0, 1), t.m_as("ms"),
filename="particlesup.gif",
text_kwargs=dict(color="white", font_size=16),
mesh_kwargs=dict(show_scalar_bar=False, color="gray", render_points_as_spheres=True),
visualize_trajectories=True,
trajectory_kwargs=dict(cmap="hot", show_scalar_bar=False, scalar_bar_args={"title": "Displacement per step [m]"}))
plotter.close()
plotter = pyvista.Plotter(off_screen=True, window_size=(500, 500))
local_functions.animate_trajectories(plotter,
np.swapaxes(r.m_as("m"), 0, 1)[:, 0:10],
t.m_as("ms"), filename="single_particle.gif",
text_kwargs=dict(color="white", font_size=16),
mesh_kwargs=dict(show_scalar_bar=False, color="gray", render_points_as_spheres=True),
visualize_trajectories=True,
trajectory_kwargs=dict(cmap="hot", scalar_bar_args={"title": "Displacement per step [m]"}))
plotter.close()
# Combine animations and display
gifs = [imageio.v3.imread(fname) for fname in ("particlesup.gif", "single_particle.gif")]
stacked_frames = [np.concatenate([f1, f2], axis=1) for f1, f2 in zip(*gifs)]
imageio.mimsave("particles.gif", stacked_frames, loop=10, duration=0.3)
b64 = base64.b64encode(open("particles.gif", 'rb').read()).decode('ascii')
display(HTML(f'<img src="data:image/gif;base64,{b64}" />'))
Fit taylor expansion and evaluate the trajectories#
[21]:
trajectory_module = cmrsim.trajectory.TaylorTrajectoryN(order=8, time_grid=t.m_as("ms"),
particle_trajectories=r.m_as("m"))
/usr/local/lib/python3.11/dist-packages/numpy/polynomial/polynomial.py:1362: RankWarning: The fit may be poorly conditioned
return pu._fit(polyvander, x, y, deg, rcond, full, w)
Evaluate positions at the reference time-points to check validity. Also compute the velocity estimation and check if it is valid
[22]:
taylor_positions, _ = trajectory_module(None, tf.constant(t.m_as("ms"), dtype=tf.float32))
taylor_positions_p, _ = trajectory_module(None, tf.constant(t.m_as("ms")+0.1, dtype=tf.float32))
taylor_velocities = (taylor_positions - taylor_positions_p).numpy() / 0.1
reference_velocities = np.gradient(r.m_as("m"), t.m_as("ms"), axis=1)
taylor_positions.numpy().shape, r.shape, taylor_velocities.shape, reference_velocities.shape
[22]:
((624, 49, 3), (624, 49, 3), (624, 49, 3), (624, 49, 3))
Visualize taylor trajectories#
[26]:
plotter = pyvista.Plotter(off_screen=True, window_size=(600, 600))
local_functions.animate_trajectories(plotter,
np.swapaxes(taylor_positions, 0, 1),
t.m_as("ms"),
filename="particlestaylor.gif",
scalars=np.swapaxes(taylor_positions-r.m_as("m"), 0, 1)*1000,
text_kwargs=dict(color="white", font_size=16),
mesh_kwargs=dict(show_scalar_bar=True, cmap="hot", render_points_as_spheres=True, scalar_bar_args={"title":"Difference to reference [mm]"}),
visualize_trajectories=False)
plotter.close()
plotter = pyvista.Plotter(off_screen=True, window_size=(600, 600))
local_functions.animate_trajectories(plotter,
np.swapaxes(taylor_positions, 0, 1) / 1000,
t.m_as("ms"),
filename="particlestaylorv.gif",
scalars=np.swapaxes(taylor_velocities-reference_velocities, 0, 1),
text_kwargs=dict(color="white", font_size=16),
mesh_kwargs=dict(show_scalar_bar=True, cmap="hot", render_points_as_spheres=True, scalar_bar_args={"title":"Difference to reference [mm/ms]"}),
visualize_trajectories=False)
plotter.close()
# Combine animations and display
gifs = [imageio.v3.imread(fname) for fname in ("particlestaylor.gif", "particlestaylorv.gif")]
stacked_frames = [np.concatenate([f1, f2], axis=1) for f1, f2 in zip(*gifs)]
imageio.mimsave("particlestay.gif", stacked_frames, loop=10, duration=0.3)
b64 = base64.b64encode(open("particlestay.gif", 'rb').read()).decode('ascii')
display(HTML(f'<img src="data:image/gif;base64,{b64}" />'))