Velocity encoded spoiled GRE with laminar flow (parallel)#
In this notebook the gradient waveforms used to instatiate the BlochOperators are evaluated in parallel, where here the multiple waveforms correspond to separate velocity encodings.
Therefore, the particle trajectories and magnetization are used for all VENCs simulateneously, which is conceptually equal to a sufficiently long pause between the acquisitions.
In contrast to the notebook demonstrating the sequential call, is this notebook one BlochOperator per TR is instantiated. Hence, the particle magnetization and position is passed from one operator to the next, in the custom loop iterating the TRs. As usual, the particles including a reseeding and pruning step to hande in- and out-flow.
The sequence is defined using CMRseq, and the meshed domain of the stenotic straight tube is provided as example resource. The particle trajectories are represented by a FlowTrajectory module, corresponding to laminar flow, which is internally called inside the Bloch simulation.
The main sections are 1. Imports 2. Sequence definition 3. Phantom setup 4. Simulation 5. Reconstruction
Imports#
[ ]:
!pip install -U cmrseq==0.23 --index-url https://gitlab.ethz.ch/api/v4/projects/30300/packages/pypi/simple
[1]:
import sys
sys.path.append("../")
sys.path.insert(0, "../../")
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
os.environ['CUDA_VISIBLE_DEVICES'] = "3"
import tensorflow as tf
gpu = tf.config.get_visible_devices("GPU")
if gpu:
tf.config.experimental.set_memory_growth(gpu[0], True)
[2]:
from IPython.display import clear_output
import pyvista
from pint import Quantity
import numpy as np
from tqdm.notebook import tqdm
import matplotlib.pyplot as plt
%matplotlib inline
import cmrsim
from cmrsim.utils import particle_properties as part_factory
import cmrseq
clear_output()
Sequence Definition#
[3]:
# Define MR-system specifications
system_specs = cmrseq.SystemSpec(max_grad=Quantity(80, "mT/m"), max_slew=Quantity(200., "mT/m/ms"),
grad_raster_time=Quantity(0.01, "ms"),
rf_raster_time=Quantity(0.01, "ms"),
adc_raster_time=Quantity(0.01, "ms"))
# Sequence Parameters
TR = Quantity(5, 'ms')
TE = Quantity(0, 'ms') # defaults to shortest
crusher_duration = Quantity(0.3, "ms")
# Velocity encoding settings:
venc_list = [Quantity(0., 'm/s'), Quantity(5.5, 'm/s'), Quantity(2., 'm/s'), Quantity(1., 'm/s')]
venc_direction_list = [np.array([0, 0, 1]), np.array([0, 0, 1]), np.array([0, 1, 0]), np.array([1, 0, 0])]
venc_duration = Quantity(1, 'ms') # Duration = 0 will result in shortest possible venc
# Define Fourier-Encoding parameters
fov = Quantity([222, 142], "mm")
matrix_size = (111, 71)
res = fov / matrix_size
print("Resolution:", res)
# Define imaging slice parameters
slice_thickness = Quantity(5, "mm")
slice_position = Quantity([0., 0., 0.], "m")
adc_duration = Quantity(1., "ms")
readout_direction = np.array([0., 0., 1.]) # readout along scanner Z, (FH)
slice_normal = np.array([1., 0., 0.])
slice_position_offset = Quantity(0, "cm")
# Define RF-exication parameters (coupled with seeding slice atm)
flip_angle = Quantity(np.pi / 12, "rad")
pulse_duration = Quantity(0.5, "ms")
RF_slice_normal = np.array([1., 0., 0.]) # Slice along scanner Y, (AP)
RF_slice_position = Quantity([0., 0., 0.], "m")
time_bandwidth_product = 4
# Define Reseeding slice parameters
seeding_slice_position = Quantity([0, 0.048, -0.045], "m")
seeding_slice_normal = np.array([1., 0., 0.])
seeding_slice_thickness = Quantity(50, "mm")
seeding_pixel_spacing = np.array((0.25e-3, 0.25e-3, 0.25e-3)) # resolution of seeding mesh, mm
seeding_slice_bb = Quantity([5, 5.1, 2], "cm")
seeding_particle_density = 10 # per 1/mm*3
# Define initial filling
initial_fill_normal = np.array([1., 0., 0.])
initial_fill_position = Quantity([-0.0005, 0., 0.], "m")
initial_fill_thickness = Quantity(50, "mm")
initial_fill_bounding_box = Quantity([25, 25, 25], "cm")
lookup_mesh_spacing = np.array((0.5e-3, 0.5e-3, 0.5e-3)) # resolution of lookup mesh, mm
Resolution: [2.0 2.0] millimeter
[5]:
rotation_matrix = cmrseq.utils.get_rotation_matrix(slice_normal, readout_direction, target_orientation='mps').T
venc_direction_list_MPS = [rotation_matrix@d for d in venc_direction_list]
crusher_area = Quantity(np.pi * 4, "rad") / system_specs.gamma_rad / res[0]
multi_list = cmrseq.contrib.pc_gre_multivenc(system_specs,
slice_thickness=slice_thickness,
flip_angle=flip_angle,
pulse_duration=pulse_duration,
time_bandwidth_product=time_bandwidth_product,
matrix_size=matrix_size,
inplane_resolution=res,
adc_duration=adc_duration,
echo_time=TE,
repetition_time=TR,
slice_position_offset=slice_position_offset,
venc_list=venc_list,
venc_direction_list=venc_direction_list_MPS,
venc_duration=venc_duration,
crusher_area=crusher_area,
crusher_duration=crusher_duration)
for sequence_list in multi_list:
for seq in sequence_list:
seq.rotate_gradients(cmrseq.utils.get_rotation_matrix(slice_normal, readout_direction, target_orientation='xyz').T)
CMRSeq Warning : Velocity Bipolar Gradient: Duration set too short
CMRSeq Warning : PC GRE Sequence: Echo time too short to be feasible, set TE to 3.1799999999999997 millisecond
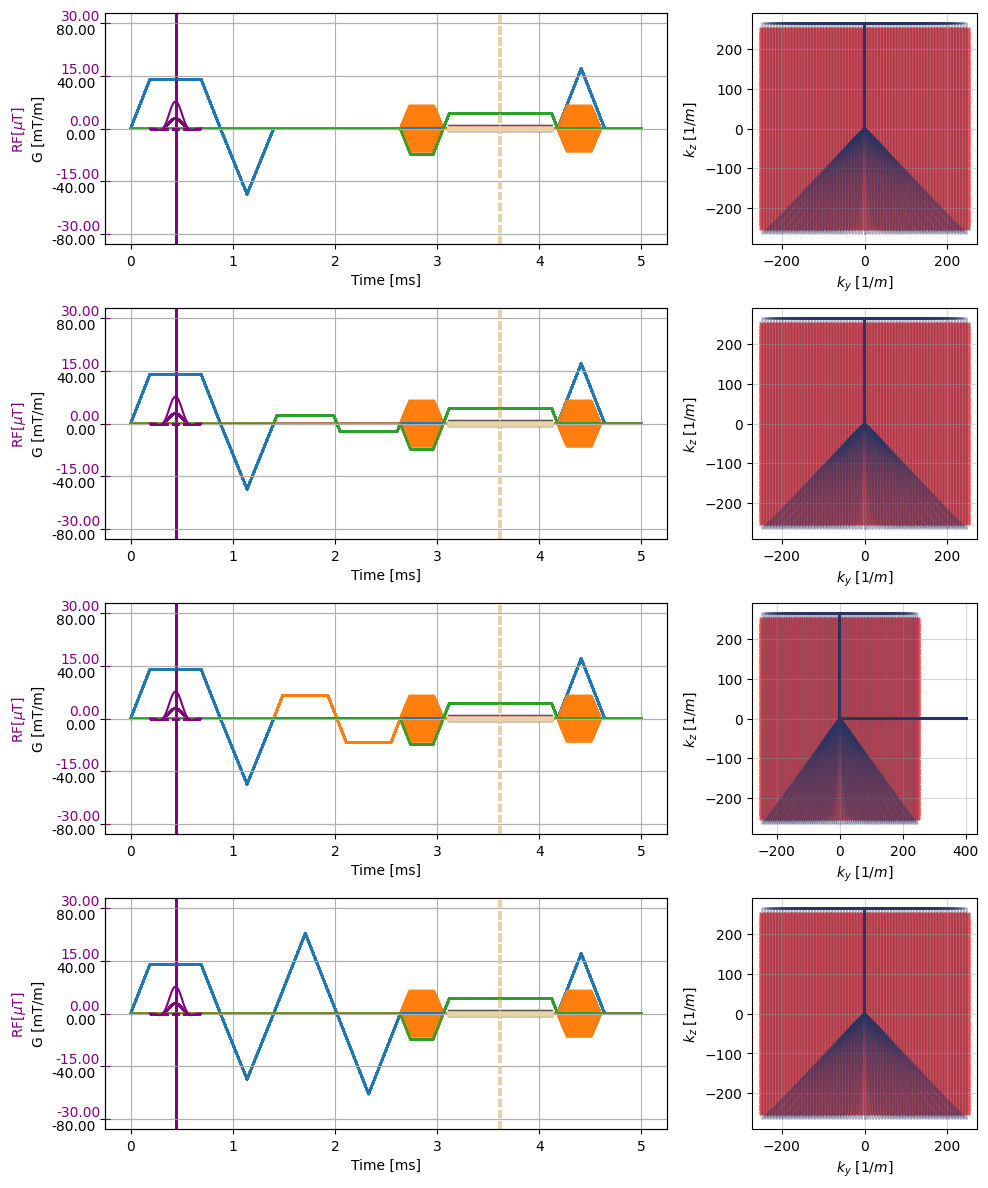
[7]:
_, k_adc, t_adc = multi_list[2][0].calculate_kspace()
r = len(multi_list)
plt.close("all")
f, ax = plt.subplots(r, 2, figsize=(10, 3*r), gridspec_kw={'width_ratios': [5, 2]})
for seq, idx in zip(multi_list, range(r)):
cmrseq.plotting.plot_sequence(seq[0], axes=ax[idx, 0], add_legend=False)
for s in seq[1:]:
cmrseq.plotting.plot_sequence(s, axes=[ax[idx, 0], ax[idx, 0], ax[idx, 0], ax[idx, 0]], format_axes=False, add_legend=False)
for s in seq:
cmrseq.plotting.plot_kspace_2d(s, ax=ax[idx, 1], k_axes=[1, 2])
f.tight_layout()
Set up phantom#
[9]:
# Load UBend Example.
mesh = pyvista.read("../example_resources/sUbend_219_9.vtu")
# The velocity [m/s] is stored in the mesh as U1. In cmrsim all definitions are done in [ms] therefore
# We add the velocity field as scaled data to the mesh
mesh["velocity"] = Quantity(mesh["U"], "m/s").m_as("m/ms")
# To match the usual geometry (Head <-> Foot along z-axis) flip the axes of the mesh
mesh.points = mesh.points[:, [2, 1, 0]]
mesh["velocity"] = mesh["velocity"][:, [2, 1, 0]]
global_mesh_offset = [0, 0.04, -0.05]
mesh.translate(global_mesh_offset, inplace=True)
mesh = mesh.cell_data_to_point_data()
# clip to restricted length to reduce mesh-size
mesh.clip(normal='-z', origin=[0., 0., -0.1], inplace=True)
# setup initial fill dict
initial_fill_dict = {'slice_normal': initial_fill_normal,
'slice_position': initial_fill_position.m_as('m'),
'slice_thickness': initial_fill_thickness.m_as('m'),
'slice_bounding_box': initial_fill_bounding_box.m_as('m')}
# Setup flow dataset with seeding mesh information
particle_creation_fncs = {"magnetization": part_factory.norm_magnetization(),
"T1": part_factory.uniform(default_value=300.),
"T2": part_factory.uniform(default_value=100.),
"M0": part_factory.uniform(default_value=1),
"omega_t2s": part_factory.t2star_lorentzian(T2p=50, prctile_cutoff=0.002)}
dataset = cmrsim.datasets.RefillingFlowDataset(mesh,
particle_creation_callables=particle_creation_fncs,
slice_position=seeding_slice_position.m_as("m"),
slice_normal=seeding_slice_normal,
slice_thickness=seeding_slice_thickness.m_as("m"),
slice_bounding_box=seeding_slice_bb.m_as("m"),
lookup_map_spacing=lookup_mesh_spacing,
seeding_vol_spacing=seeding_pixel_spacing,
field_list=[("velocity", 3), ])
mesh
Updated Mesh
Updated Slice Position
[9]:
| Header | Data Arrays | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
Simulation#
Define Bloch operators#
[15]:
# Define off-resonsance and T2* modules
T2s = cmrsim.bloch.submodules.T2starDistributionModel()
submodules = [T2s, ]
[16]:
time_list = []
rf_list = []
grad_list = []
adc_list = []
for sequence_list in multi_list:
time, rf_grid, grad_grid, adc_on_grid = [np.stack(v) for v in cmrseq.utils.grid_sequence_list(sequence_list, force_uniform_grid=False)]
time_list.append(time)
rf_list.append(rf_grid)
grad_list.append(grad_grid)
adc_list.append(adc_on_grid)
time_ref = time_list[0][0]
adc_all = np.stack(adc_list, axis=1)
grad_all = np.stack(grad_list, axis=1)
rf_all = np.stack(rf_list, axis=1)
bloch_list = []
for tr in range(grad_all.shape[0]):
bloch_list.append(cmrsim.bloch.GeneralBlochOperator(name="rf", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
time_grid=time_ref, gradient_waveforms=grad_all[tr],
rf_waveforms=rf_all[tr], adc_events=adc_all[tr], device="GPU:0",
submodules=submodules))
Run#
The loop over TRs running the the VENCS in parallel is implemented in a corresponding simulation template for convenience. It also includes the display of a infomative progress bar.
[17]:
from cmrsim.simulation_templates.flow import FlowSimulationMultiVenc
simulator = FlowSimulationMultiVenc(dataset, prep_modules=[], readout_modules=bloch_list)
r, m = simulator(particle_density=seeding_particle_density, dropping_distance=0.2, averages=1,
reseed_threshold=1, use_inital_filling=True,
initial_filling_dict=initial_fill_dict, return_final_magnetization=True)
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
I0000 00:00:1712913156.979724 2980859 device_compiler.h:186] Compiled cluster using XLA! This line is logged at most once for the lifetime of the process.
WARNING:tensorflow:5 out of the last 5 calls to <function GeneralBlochOperator._call_core at 0x7f55ede287c0> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has reduce_retracing=True option that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
WARNING:tensorflow:6 out of the last 6 calls to <function GeneralBlochOperator._call_core at 0x7f55ede2ade0> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has reduce_retracing=True option that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
Reconstruction#
[20]:
signal_list = []
for bloch_mod in bloch_list:
signal_list.append(tf.stack(bloch_mod.time_signal_acc, axis=0).numpy())
signals = np.stack(signal_list,axis=1)
image_list = []
kspace_list = []
for ind in range(np.shape(signals)[0]):
time_signal = np.squeeze(signals[ind, :, :])
n = (np.random.normal(0, 1, time_signal.shape)) * 100
n = n*np.exp(1j*np.random.normal(0, 1, time_signal.shape))
time_signal = time_signal + n
center_index = np.array(matrix_size) // 2 + np.mod(matrix_size, 2)
image_list.append(tf.signal.fftshift(tf.signal.ifft2d(tf.signal.ifftshift(time_signal, axes=(0, 1))), axes=(0,1)).numpy())
kspace_list.append(time_signal)
[32]:
# Show images
fig, axes = plt.subplots(len(venc_list), 3, figsize=(8, 2*len(venc_list)))
for image, k_space, idx in zip(image_list, kspace_list, range(len(venc_list))):
axes[idx, 0].imshow(np.log(np.abs(np.squeeze(k_space))), cmap="gray")
axes[idx, 1].imshow(np.abs(np.squeeze(image)), cmap="gray", vmin=0, vmax=np.percentile(np.abs(np.squeeze(image)), 99.9))
axes[idx, 2].imshow(np.angle(np.squeeze(image)), cmap="RdBu", vmin=-np.pi, vmax=np.pi)
[_.axis("off") for _ in axes.flatten()]
[_.set_title(t) for _, t in zip(axes[0, :], ["k-space", "Magnitude", "Phase"])]
fig.tight_layout()
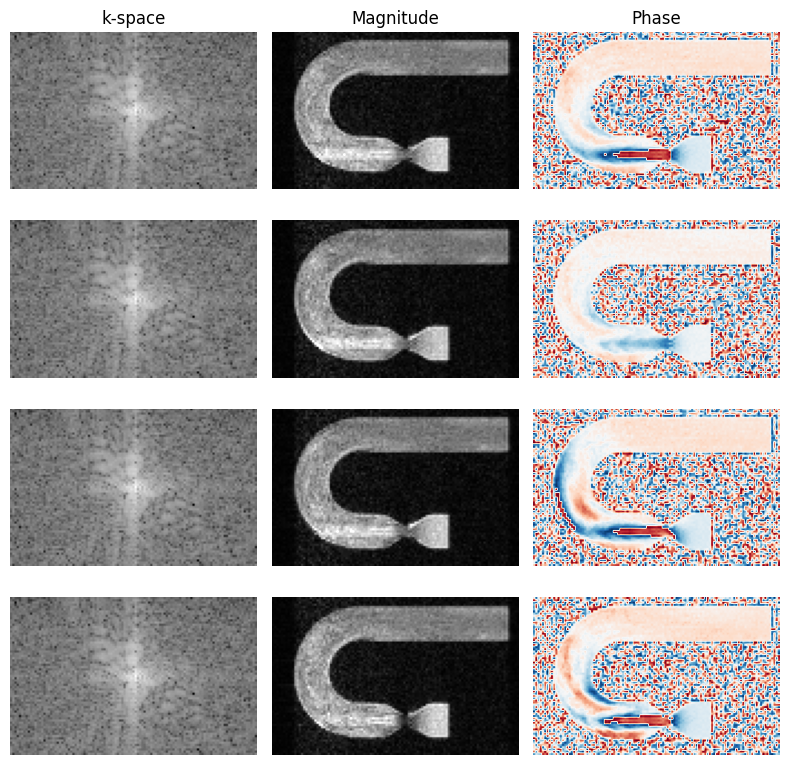
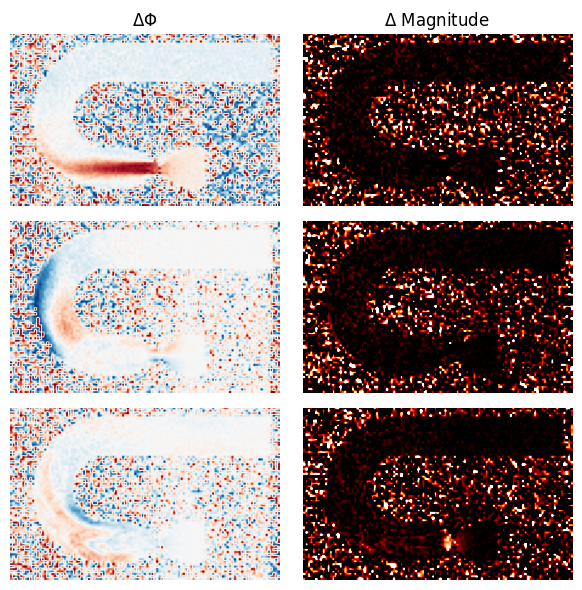
[39]:
# show phase and magnitude difference to references
fig, axes = plt.subplots(len(venc_list)-1, 2, figsize=(6, 2*(len(venc_list)-1)))
ref_im = image_list[0]
mask = np.abs(ref_im) > 0
axes = axes.flatten()
for image, idx in zip(image_list[1:], range(len(venc_list) -1)):
axes[2*idx].imshow(mask*np.mod(np.angle(np.squeeze(image)) - np.angle(np.squeeze(ref_im)) + np.pi, 2 * np.pi) - np.pi, cmap="RdBu", vmin=-np.pi, vmax=np.pi)
axes[2*idx+1].imshow(mask*np.log(np.abs(np.squeeze(ref_im))/np.abs(np.squeeze(image))), cmap="gist_heat", vmin=0, vmax=1)
[_.axis("off") for _ in axes.flatten()]
[_.set_title(t) for _, t in zip(axes[:], [r"$\Delta \Phi$", r"$\Delta$ Magnitude"])]
fig.tight_layout()
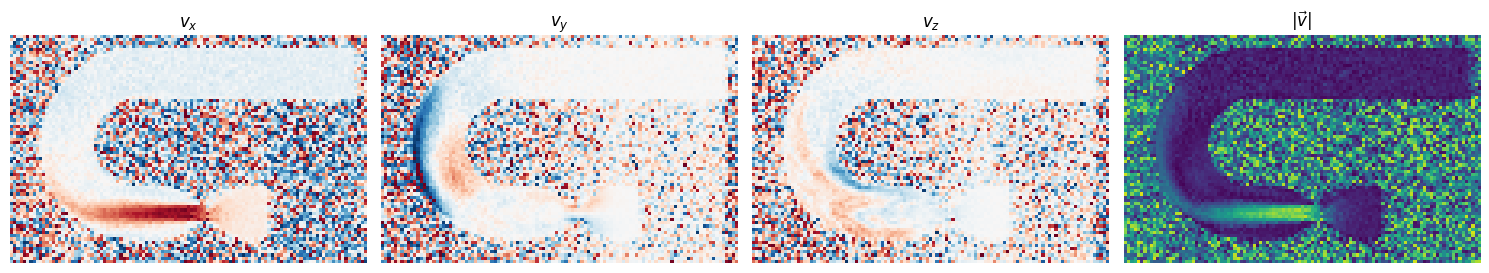
[40]:
fig, axes = plt.subplots(1, len(venc_list), figsize=(5*(len(venc_list)-1), 6))
axes = axes.T
ref_im = image_list[0]
mask = np.abs(ref_im) > 0
axes = axes.flatten()
vlist = []
for image, idx in zip(image_list[1:], range(len(venc_list)-1)):
venc_cur = venc_list[idx+1].m_as("cm/s")
v = (np.mod(np.angle(np.squeeze(image))-np.angle(np.squeeze(ref_im)) + np.pi, 2 * np.pi) - np.pi)/np.pi * venc_cur
vlist.append(v)
axes[idx].imshow(v, cmap="RdBu", vmin=-venc_cur, vmax=venc_cur)
axes[-1].imshow(np.sqrt(np.sum(np.asarray(vlist)**2, axis=0)), cmap="viridis", vmin=0, vmax=600)
[_.set_title(t) for _, t in zip(axes[:], ["$v_x$", "$v_y$", "$v_z$", r"$|\vec{v}|$"])]
[_.axis("off") for _ in axes.flatten()]
fig.tight_layout()