Static Phantom with off-resonance#
In this notebook, a simulation of a balanced steady state free precession with a spherical static phantom with air inclusions is performed. To do so the following steps are performed:
Define the Analytic Simulator
Define a 3D spherical phantom
Run simulation
Reconstruct images
Import#
[1]:
!pip install cmrseq --index-url https://gitlab.ethz.ch/api/v4/projects/30300/packages/pypi/simple
[2]:
import sys
sys.path.append("../")
sys.path.insert(0, "../../")
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
os.environ['CUDA_VISIBLE_DEVICES'] = "3"
import tensorflow as tf
gpu = tf.config.get_visible_devices("GPU")
if gpu:
tf.config.experimental.set_memory_growth(gpu[0], True)
[3]:
import base64
from IPython.display import display, HTML, clear_output
# 3rd Party dependencies
from pint import Quantity
import pyvista
import numpy as np
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
import imageio
%matplotlib inline
# Project library cmrsim
import cmrseq
import cmrsim
import local_functions
1. Define Analytical Simulator#
[6]:
from typing import Tuple
class BSSFPSimulator(cmrsim.analytic.simulation.AnalyticSimulation):
def build(self, fov: Tuple[float, float], matrix_size: Tuple[int, int], flip_angle: float, TE: float, TR: Quantity = Quantity(0, "ms"),
slice_thickness: Quantity = Quantity(10, "mm"), adc_duration: Quantity = Quantity(2, "ms"),
pulse_duration: Quantity = Quantity(0.5, "ms"), repetition_time=Quantity(0, "ms")):
n_kx, n_ky = matrix_size
# Encoding definition
inplane_resolution = Quantity(fov, "m") / matrix_size
sequence_list = cmrseq.seqdefs.sequences.balanced_ssfp(cmrseq.SystemSpec(), matrix_size, inplane_resolution,
slice_thickness=slice_thickness, adc_duration=adc_duration,
repetition_time=TR,
flip_angle=Quantity(flip_angle, "degree"),
pulse_duration=pulse_duration,
dummy_shots=0)
encoding_module = cmrsim.analytic.encoding.GenericEncoding.from_cmrseq(name="bssfp_readout",
sequence=sequence_list[1:],
absolute_noise_std=0., device="GPU:0")
# # Signal module construction
sequence_module = cmrsim.analytic.contrast.BSSFP(flip_angle=np.array([flip_angle, ]), echo_time=[TE, ],
repetition_time=[float(TR.m_as("ms")), ], expand_repetitions=False,
# device="GPU:0"
)
# Forward model composition
forward_model = cmrsim.analytic.CompositeSignalModel(sequence_module)
return forward_model, encoding_module, None
[7]:
matrix_size = np.array([151, 151])
fov = Quantity([22, 22], "cm")
simulator = BSSFPSimulator(build_kwargs=dict(fov=fov.m_as("m").astype(np.float32),
matrix_size=matrix_size, flip_angle=35,
TE=6))
CMRSeq Warning : bSSFP Sequence: Repetition time too short to be feasible, set TR to 4.279999999999999 millisecond
2. Define Phantom#
Create 3D object#
[8]:
phantom = local_functions.create_spherical_phantom(dimensions=(61, 61, 61), spacing=(0.004, 0.004, 0.004))
clear_output()
# Create field to signal if regular mesh-points are within original unstructured grid
phantom["in_mesh"] = np.ones(phantom.points.shape[0])
Instantiate a RegularGridDataset#
Also assign MRI relevant material properties and comput the off-resonance distribution
[9]:
# Create regular grids
dataset = cmrsim.datasets.RegularGridDataset.from_unstructured_grid(phantom, pixel_spacing=Quantity([0.5, 0.5, 0.5], "mm"), padding=Quantity([5, 5, 5], "cm"))
# Assign properties (everything not 'in-mesh' is assumed to be air)
dataset.mesh["chi"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]) * (-9.05), np.ones_like(dataset.mesh["in_mesh"]) * 0.36) * 1e-6 # susceptibilty in ppm
dataset.mesh["M0"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]), np.zeros_like(dataset.mesh["in_mesh"])) # density in percent
dataset.mesh["T1"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]) * 1000, np.zeros_like(dataset.mesh["in_mesh"])) # time in ms
dataset.mesh["T2"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]) * 300, np.zeros_like(dataset.mesh["in_mesh"])) # time in ms
dataset.mesh["T2star"] = np.where(dataset.mesh["in_mesh"], np.ones_like(dataset.mesh["in_mesh"]) * 100, np.zeros_like(dataset.mesh["in_mesh"])) # time in ms
[10]:
dataset.compute_offresonance(b0=Quantity(1.5, "T"), susceptibility_key="chi");
CMRSeq RuntimeWarning: /scratch/jweine/cmrsim/notebooks/analytic_simulation/../../cmrsim/datasets/_regular_grid.py:192 : invalid value encountered in divide
CMRSeq UnitStrippedWarning: /usr/local/lib/python3.11/dist-packages/pyvista/core/dataset.py:1975 : The unit of the quantity is stripped when downcasting to ndarray.
Select a slice#
The slice coordinates are transformed into MPS coordinates as visualized below. The selected slice-mesh is wrapped again into a RegularGridDataset to subsequently extract the non-trivial material points as dictionary of arrays.
[11]:
# Define imaging slice parameters
slice_thickness = Quantity(5, "mm")
readout_direction = np.array([0., 1., 0.])
slice_normal = np.array([1., 0., 0.])
slice_position_offset = Quantity(0., "cm")
slice_mesh = dataset.select_slice(slice_normal=slice_normal, spacing=Quantity((0.25, 0.25, 3.), "mm"),
slice_position=slice_normal*slice_position_offset,
field_of_view=Quantity([22, 22, slice_thickness.m_as("cm")], "cm"),
readout_direction=readout_direction, in_mps=True)
slice_dataset = cmrsim.datasets.RegularGridDataset(slice_mesh)
phantom_dict = slice_dataset.get_phantom_def(filter_by="in_mesh", keys=["M0", "T1", "T2", "offres"],
perturb_positions_std=Quantity(0.005, "mm").m_as("m"))
CMRSeq Warning : Optimal rotation is not uniquely or poorly defined for the given sets of vectors.
Plot 3D phantom with offresonance map
[12]:
pyvista.close_all()
pyvista.start_xvfb()
plotter = pyvista.Plotter(notebook=False, off_screen=True, window_size=(600, 400))
plotter.add_mesh(dataset.mesh.threshold(0.1, "M0"), scalars="offres", opacity=0.65,
cmap="twilight", scalar_bar_args=dict(title="Offresonance (T)"))
box = local_functions.transformed_box(np.eye(3, 3), slice_normal, readout_direction, slice_position_offset*slice_normal,
extend=Quantity([*fov.m_as("m"), slice_thickness.m_as("m")], "m"))
plotter.add_mesh(box)
local_functions.add_custom_axes(plotter)
plotter.screenshot("polydata_temp0.png")
plotter.close()
plotter = pyvista.Plotter(notebook=False, off_screen=True, window_size=(600, 400))
plotter.add_mesh(pyvista.PolyData(phantom_dict["r_vectors"]), scalars=phantom_dict["offres"],
opacity=0.65, cmap="twilight", scalar_bar_args=dict(title="Offresonance (T)"))
local_functions.add_custom_axes(plotter)
plotter.screenshot("polydata_temp1.png")
img = np.concatenate([imageio.v3.imread("polydata_temp0.png"), imageio.v3.imread("polydata_temp1.png")], axis=1)
imageio.v3.imwrite("polydata_temp.png", img)
b64 = base64.b64encode(open("polydata_temp.png", 'rb').read()).decode('ascii')
display(HTML(f'<img src="data:image/gif;base64,{b64}" />'))
CMRSeq Warning : Optimal rotation is not uniquely or poorly defined for the given sets of vectors.
3. Perfom Simulation#
Create Bloch-dataset for batched data stream#
[13]:
properties = {"M0": phantom_dict["M0"].astype(np.complex64),
"T1": phantom_dict["T1"].astype(np.float32),
"T2": phantom_dict["T2"].astype(np.float32),
"T2star": phantom_dict["T2"].astype(np.float32),
"off_res": (phantom_dict["offres"]).astype(np.float32) * Quantity(42.5, "MHz/T").m_as("1/ms/mT") * 2 * np.pi,
"r_vectors": phantom_dict["r_vectors"].astype(np.float32)
}
input_dataset = cmrsim.datasets.AnalyticDataset(properties, filter_inputs=True, expand_dimension=True)
print(properties["M0"].shape)
(473540,)
Call simulator object#
[14]:
k_space = simulator(input_dataset, voxel_batch_size=int(1e5)).numpy().reshape(matrix_size)
Run Simulation: |XXXXXXXXXXXXXXX|473540/473540 |XXXXXXXXXXXXXXX|151/151
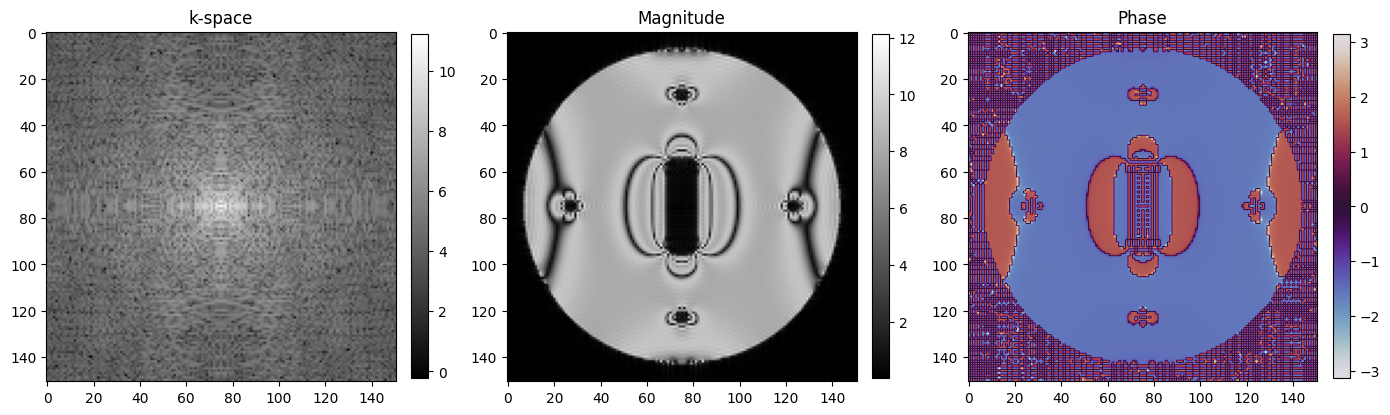
4. Reconstruct images#
[15]:
image = tf.signal.fftshift(tf.signal.ifft2d(tf.signal.ifftshift(k_space, axes=(0, 1))), axes=(0, 1)).numpy()
[16]:
# Show images
fig, axes = plt.subplots(1, 3, figsize=(14, 5))
kspace_plot = axes[0].imshow(np.log(np.abs(np.squeeze(k_space))), cmap="gray")
fig.colorbar(kspace_plot, ax=axes[0], fraction=0.045, pad=0.04)
abs_plot = axes[1].imshow(np.abs(np.squeeze(image)), cmap="gray")
fig.colorbar(abs_plot, ax=axes[1], fraction=0.045, pad=0.04)
phase_plot = axes[2].imshow(np.angle(np.squeeze(image)), cmap="twilight", vmin=-np.pi, vmax=np.pi)
fig.colorbar(phase_plot, ax=axes[2], fraction=0.045, pad=0.04)
# [_.axis("off") for _ in axes]
[_.set_title(t) for _, t in zip(axes, ["k-space", "Magnitude", "Phase"])]
fig.tight_layout()