LV Contraction and breathing motion with bSSFP#
This notebook contains the code to reproduce the introductory example described in the CMRseq publication.
At the end of this notebook, the non-executable code excerpt included in the paper is appended.
Imports#
[ ]:
!pip install cmrseq --index-url https://gitlab.ethz.ch/api/v4/projects/30300/packages/pypi/simple
[1]:
import sys
sys.path.append("../")
sys.path.insert(0, "../../")
sys.path.insert(0, "../../../cmrseq")
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
os.environ['CUDA_VISIBLE_DEVICES'] = "3"
import tensorflow as tf
gpu = tf.config.get_visible_devices("GPU")
if gpu:
tf.config.experimental.set_memory_growth(gpu[0], True)
[11]:
from copy import deepcopy
import base64
from IPython.display import display, HTML, clear_output, Image
import pyvista
from pint import Quantity
import numpy as np
from tqdm.notebook import tqdm
import matplotlib.pyplot as plt
%matplotlib inline
import cmrsim
import cmrseq
import local_functions
clear_output()
Load cardiac mesh#
The full resource can be downloaded and unzipped into the local folder “test_data” as follows
[3]:
!wget https://gitlab.ethz.ch/ibt-cmr/modeling/cmr-random-diffmaps/-/archive/main/cmr-random-diffmaps-main.zip?path=notebooks/example_data -O test_data.zip
!unzip test_data.zip
!rm test_data.zip -r
!mv cmr-random-diffmaps-main-notebooks-example_data/notebooks/example_data test_data
!rm cmr-random-diffmaps-main-notebooks-example_data -r
clear_output()
Now load the meshes (from vtk files) and the timings as defined in the text file PV_loop_reordered.txt
[4]:
files = [f'test_data/example_mesh/Displacements/Displ{i:03}.vtk' for i in range(0, 200, 1)]
timing = Quantity(np.loadtxt("test_data/example_mesh/PV_loop_reordered.txt")[:, 1], "ms")
original_mesh = cmrsim.datasets.CardiacMeshDataset.from_list_of_meshes(files, timing[:len(files)],
mesh_numbers=(4, 40, 30),
time_precision_ms=3)
Loading Files... : 100%|██████████| 199/199 [00:03<00:00, 51.88it/s]
Refine mesh#
To reduce discretization artifacts, the mesh is refined at each snapshot. As intermediate result, the single vtk is saved.
[27]:
refined_mesh = original_mesh.refine(longitudinal_factor=5, circumferential_factor=4, radial_factor=5)
refined_mesh.mesh.save(f"refined_mesh_{tuple(refined_mesh.mesh_numbers[i] for i in (2, 0, 1))}.vtk")
Save cell volumes#
First the previously refined and saved mesh is loaded, and subsequently the cell sizes are converted to factors describing the available equlibrium magnetization per mesh node.
[30]:
fn = "refined_mesh_(16, 161, 150).vtk"
mesh_numbers = [int(s) for s in fn.replace("refined_mesh_(", "").replace(").vtk", "").split(",")]
refined_mesh = cmrsim.datasets.CardiacMeshDataset.from_single_vtk(f"{fn}", time_precision_ms=3, mesh_numbers=mesh_numbers)
[31]:
ref_time, ref_pos = refined_mesh.get_trajectories(start=Quantity(0, "ms"), end=Quantity(refined_mesh.timing[-1] + 1, "ms"))
connectivity = refined_mesh.mesh.cells_dict[10].reshape(-1, 4)
proton_density_weights = np.stack([cmrsim.datasets.CardiacMeshDataset.evaluate_cellsize(ref_pos[:, i].m_as("m"), connectivity)[2]
for i in tqdm(range(ref_time.shape[0]), desc="Calculating cell volumes", leave=False)])[..., np.newaxis]
np.save("pd.npy", proton_density_weights)
Inititalize Trajectory modules#
The following three modules are instantiated: 1. M0 - interpolation 2. Particle position due to contraction 3. Breathing wrapper
[32]:
ref_time, ref_pos = refined_mesh.get_trajectories(start=Quantity(0, "ms"), end=Quantity(refined_mesh.timing[-1] + 1, "ms"))
proton_density_weights = np.load("pd.npy")
batch_size = 400_000
proton_density_module = cmrsim.trajectory.PODTrajectoryModule(ref_time.m_as("ms"), proton_density_weights.transpose(1, 0, 2),
n_modes=10, poly_order=10, batch_size=batch_size, is_periodic=True)
contraction_module = cmrsim.trajectory.PODTrajectoryModule(ref_time.m_as("ms"), ref_pos.m_as("m"),
n_modes=10, poly_order=10,
batch_size=batch_size, is_periodic=True)
breathing_module = cmrsim.trajectory.SimpleBreathingMotionModule.from_sinosoidal_motion(sub_trajectory=contraction_module,
breathing_period=Quantity(7, "s"),
breathing_direction=np.array([1, 1, 3]),
breathing_amplitude=Quantity(2.5, "cm"))
proton_density_weights.shape, proton_density_weights.dtype, ref_time.shape, ref_time.dtype
(3,)
CMRSeq RuntimeWarning: /usr/local/lib/python3.11/dist-packages/numpy/polynomial/polyutils.py:660 : overflow encountered in square
CMRSeq RuntimeWarning: /usr/local/lib/python3.11/dist-packages/numpy/core/_methods.py:49 : overflow encountered in reduce
CMRSeq RankWarning: /usr/local/lib/python3.11/dist-packages/numpy/polynomial/polynomial.py:1362 : The fit may be poorly conditioned
(3,)
[32]:
((198, 386416, 1), dtype('float64'), (198,), dtype('float64'))
Define Sequence#
The cmrseq package is used for sequence definition.
[33]:
# Define MR-system specifications
system_specs = cmrseq.SystemSpec(max_grad=Quantity(50, "mT/m"), max_slew=Quantity(200., "mT/m/ms"),
grad_raster_time=Quantity(0.01, "ms"),
rf_raster_time=Quantity(0.01, "ms"),
adc_raster_time=Quantity(0.01, "ms"),
b0=Quantity(1.5, "T")
)
# Define Fourier-Encoding parameters
fov = Quantity([15, 15], "cm")
matrix_size = (101, 101)
n_dummy = 81
seq_list = cmrseq.seqdefs.sequences.balanced_ssfp(system_specs,
matrix_size,
repetition_time=Quantity(0., "ms"),
inplane_resolution=fov / matrix_size,
slice_thickness=Quantity(5, "mm"),
adc_duration=Quantity(2., "ms"),
flip_angle=Quantity(np.pi/4, "rad"),
pulse_duration=Quantity(0.6, "ms"),
slice_position_offset=Quantity(0., "cm"),
dummy_shots=n_dummy)
CMRSeq Warning : bSSFP Sequence: Repetition time too short to be feasible, set TR to 4.16 millisecond
[12]:
_, k_adc, t_adc = seq_list[n_dummy + 51].calculate_kspace()
full_seq = seq_list[0].copy()
full_seq.extend(seq_list[1:])
plt.close("all")
f = plt.figure(constrained_layout=True, figsize=(15, 8))
axes = f.subplot_mosaic("AAB;CCC")
cmrseq.plotting.plot_sequence(seq_list[0], axes=axes["A"], format_axes=True, add_legend=False)
for s in tqdm(seq_list[n_dummy:]):
cmrseq.plotting.plot_sequence(s, axes=[f.axes[-1], axes["A"], axes["A"], axes["A"]], format_axes=False)
for s in tqdm(seq_list[n_dummy:]):
cmrseq.plotting.plot_kspace_2d(s, ax=axes["B"], k_axes=[0, 1])
cmrseq.plotting.plot_sequence(full_seq, axes=axes["C"])
f.savefig("sequence_plot.png", dpi=400)
clear_output()
display(Image("sequence_plot.png"))
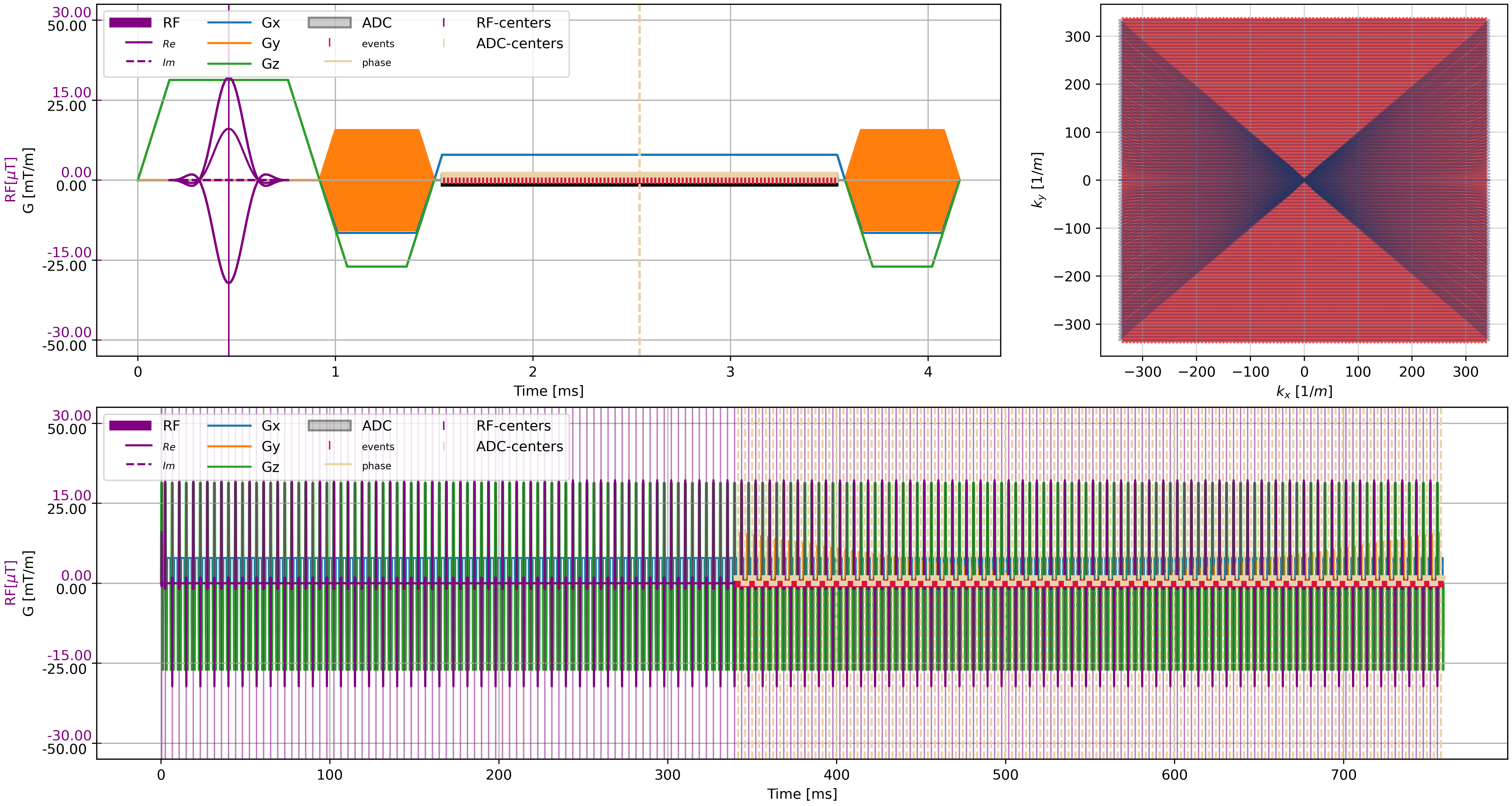
Simulation#
Initialize Bloch operators#
Again cmrseq is used to create the gridded version of all waveforms.
[34]:
dummy_sequence = seq_list[0].copy()
dummy_sequence.extend(seq_list[1:n_dummy+1])
time_dummy, rf_grid_dummy, grad_grid_dummy, _ = [np.stack(v) for v in cmrseq.utils.grid_sequence_list([dummy_sequence, ])]
time, rf_grid, grad_grid, adc_on_grid = [np.stack(v) for v in cmrseq.utils.grid_sequence_list(tqdm(seq_list[n_dummy+1:]))]
print(time_dummy.shape, rf_grid_dummy.shape, grad_grid_dummy.shape)
print(time.shape, rf_grid.shape, grad_grid.shape, adc_on_grid.shape)
# Construct BlochOperators and pass offresonance module:
module_dummyshots = cmrsim.bloch.GeneralBlochOperator(name="dummy_shots", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
time_grid=time_dummy[0],
gradient_waveforms=grad_grid_dummy,
rf_waveforms=rf_grid_dummy,
device="GPU:0")
module_acquisition = cmrsim.bloch.GeneralBlochOperator(name="acquisition", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
time_grid=time[0],
gradient_waveforms=grad_grid,
rf_waveforms=rf_grid,
adc_events=adc_on_grid,
device="GPU:0")
Extending Sequence: 100%|██████████| 81/81 [00:00<00:00, 241.05it/s]
(1, 33905) (1, 33905) (1, 33905, 3)
(101, 517) (101, 517) (101, 517, 3) (101, 517, 2)
Setup input data#
[35]:
end_dummy = time_dummy[0, -1]
start_of_trs = (np.array([-end_dummy, 0, *np.cumsum(time[:, -1]).tolist()]) + end_dummy).astype(np.float32)
M0_initial = np.concatenate([proton_density_module(None, start_of_trs[0], batch_index=b)[0][:, 0, 0] for b in range(3)], axis=0)
properties = {"M0": M0_initial,
"T1": np.ones_like(M0_initial) * 1000,
"T2": np.ones_like(M0_initial) * 200,
"magnetization": cmrsim.utils.particle_properties.norm_magnetization()(M0_initial.shape[0]),
"initial_position": ref_pos[:, 0].m_as("m").astype(np.float32)
}
input_dataset = cmrsim.datasets.BlochDataset(properties, filter_inputs=False)
print(properties["M0"].shape, properties["initial_position"].shape)
(386416,) (386416, 3)
Run#
Now run the simulation for all cases (no motion, contraction only, and respiratory+contraction), by using the corresponding trajectory module. The call to the Bloch operators is identical otherwise (two inner loops).
[37]:
tf.config.run_functions_eagerly(False)
print(f"Total time-steps per TR: {time.shape[1]}")
batch_size_int = int(contraction_module.batch_size.read_value())
# Loop over motion - cases
pbar_motion = tqdm(zip(["static", "contraction", "breathing"],
[cmrsim.trajectory.StaticParticlesTrajectory(),
contraction_module, breathing_module]),
desc="Loop over Motion cases", leave=False, total=3)
for name, tr_mod in pbar_motion:
# Reset signal accumulation
module_acquisition.reset()
# Loop over dataset batches
for batch_index, batch in tqdm(input_dataset(batchsize=batch_size_int).enumerate()):
# Reset internal time of trajectory
breathing_module.current_time_ms.assign(0.)
contraction_module._taylor_module.current_time_ms.assign(0.)
m_init = batch.pop("magnetization")
initial_position = batch.pop("initial_position")
# Set M0 according to cell volume at TR
m0 = proton_density_module(initial_position, start_of_trs[0],
batch_index=tf.cast(batch_index, tf.int32))[0][:, 0, 0]
batch["M0"] = m0
contraction_module.current_batch_idx.assign(tf.cast(batch_index, dtype=tf.int32))
m, r = module_dummyshots(initial_position=initial_position, magnetization=m_init,
**batch, trajectory_module=tr_mod)
pbar = tqdm(range(time.shape[0]), desc="Iterating TRs", leave=False)
for tr_index in pbar:
pbar.set_postfix_str(f"Current simulation time: {contraction_module.current_time_ms.read_value(): 1.3f}")
m0 = proton_density_module(initial_position, start_of_trs[tr_index], batch_index=tf.cast(batch_index, tf.int32))[0][:, 0, 0]
batch["M0"] = m0
m, r = module_acquisition(initial_position=r, magnetization=m, repetition_index=tr_index,
**batch, trajectory_module=tr_mod)
# Reconstruct image and save results
time_signal = tf.stack(module_acquisition.time_signal_acc, axis=0).numpy().reshape(matrix_size[::-1], order="F")
time_signal += tf.complex(*[tf.random.normal(shape=time_signal.shape, stddev=30) for i in range(2)])
image = tf.signal.fftshift(tf.signal.ifft2d(tf.roll(tf.signal.ifftshift(time_signal, axes=(0, 1)), -1, axis=1)), axes=(0, 1)).numpy()
np.savez(f"{name}_result.npz", time_signal=time_signal.numpy(), image=image)
Total time-steps per TR: 517
Create figure for publication#
[21]:
start, end = 0., contraction_module._taylor_module.current_time_ms.read_value().numpy()
pos, _ = breathing_module(ref_pos.m, np.array([0., 750.], dtype=np.float32))
temp_mesh_start = pyvista.UnstructuredGrid(refined_mesh.mesh.cells_dict, pos[:, 0].numpy())
temp_mesh_end = pyvista.UnstructuredGrid(refined_mesh.mesh.cells_dict, pos[:, 1].numpy())
coms = np.mean(pos, axis=0)
sphere1 = pyvista.Sphere(radius=0.001, center=coms[0])
sphere2 = pyvista.Sphere(radius=0.001, center=coms[1])
pyvista.close_all()
pyvista.start_xvfb()
pyvista.set_jupyter_backend("static")
plotter = pyvista.Plotter(off_screen=True, window_size=(800, 800))
plotter.add_mesh(temp_mesh_start, show_scalar_bar=False, color="lightblue", opacity=0.2)
plotter.add_mesh(sphere1, color="lightblue")
plotter.add_mesh(temp_mesh_end, show_scalar_bar=False, color="lightgreen", opacity=0.2)
plotter.add_mesh(sphere2, color="lightgreen")
plotter.add_arrows(coms[0:1], coms[1:2] - coms[:1], color="red")
box2 = local_functions.transformed_box(reference_basis=np.eye(3, 3), slice_normal=np.array([0., 0., 1.]),
readout_direction=np.array([1., 0., 0.]),
slice_position=Quantity([0., 0., -2.], "cm"),
extend=Quantity([10., 10., 1.], "cm"))
plotter.add_mesh(box2, color="black", opacity=0.3, show_edges=True)
plotter.camera_position = "xz"
plotter.screenshot("range_of_motion.png", window_size=(1000, 1000));
plotter.close()
CMRSeq Warning : Optimal rotation is not uniquely or poorly defined for the given sets of vectors.
[38]:
import imageio
plt.rcParams["font.family"] = "serif"
figure, axes = plt.subplots(1, 4, constrained_layout=True, figsize=(15, 5))
[ax.set_title(t, fontsize=18) for ax, t in zip(axes, ["Range of Motion", "Breathing + Contraction", "Contraction", "Static"])]
[ax.axis("off") for ax in axes]
axes[0].imshow(imageio.v3.imread("range_of_motion.png"))
axes[1].imshow(np.abs(np.load("breathing_result.npz")["image"]), cmap="gray")
axes[2].imshow(np.abs(np.load("contraction_result.npz")["image"]), cmap="gray")
axes[3].imshow(np.abs(np.load("static_result.npz")["image"]), cmap="gray")
[ax.text(0.05, 0.95, f"{t})", horizontalalignment='left', verticalalignment='top',
transform=ax.transAxes, fontsize=18, fontweight="bold", color=c)
for ax, t, c in zip(axes, "abcd", "kwww")]
figure.savefig("movemesh_bssfp.png", dpi=300)
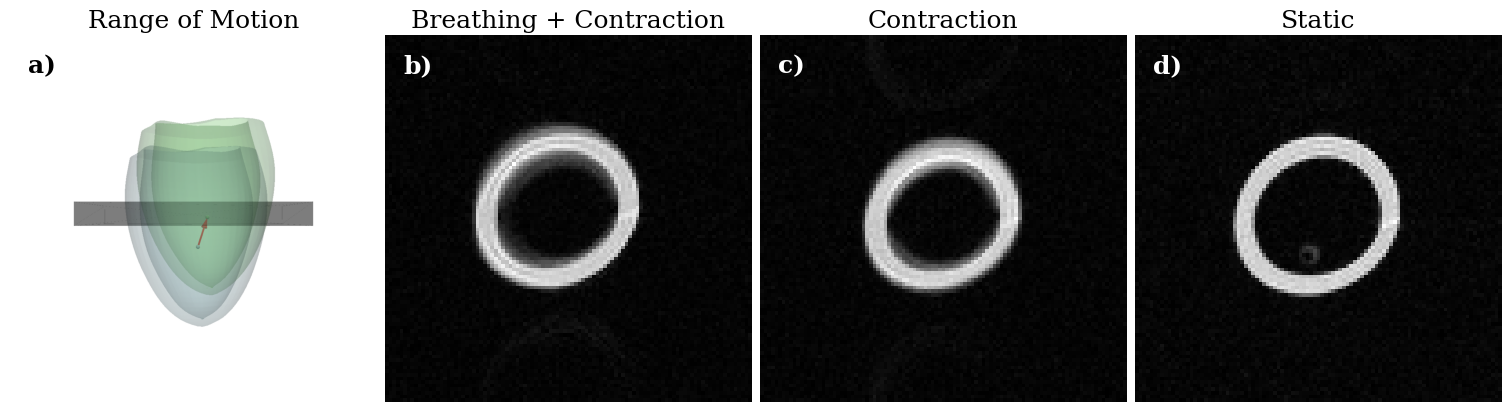
# Python Code corresponding to Pseudo Code in Figure 2#
The code shown below only contains the lines, that are relevant for the statements contained in the pseudo code in Figure 2. Therefore it is not executable as is. To access the complete, runnable example code please visit the repository and/or the documentation webpage via: https://cmr.ethz.ch/research/software.html.
Preparation / Load resources#
[ ]:
files = [f'{resource_path}/Displ{i:03}.vtk' for i in range(0, 228, 1)]
timing = Quantity(np.loadtxt(f"{resource_path}/snapshot_timings.txt"), "ms")
refine_module = cmrsim.datasets.CardiacMeshDataset.from_list_of_meshes(files, timing, ...)
refined_mesh = refine_module.refine(longitudinal_factor=5, circumferential_factor=4, radial_factor=5)
ref_times, ref_pos = refined_mesh.get_trajectories(...)
pd_weights_t = [cmrsim.datasets.CardiacMeshDataset.evaluate_cellsize(...) for t in ref_times]
proton_density_weights = np.stack(pd_weights_t)
Initialize Modules#
[ ]:
batch_size=200_000
proton_density_module = cmrsim.trajectory.PODTrajectoryModule(ref_time.m_as("ms"), proton_density_weights,
n_modes=10, poly_order=10,
batch_size=batch_size, is_periodic=True)
contraction_module = cmrsim.trajectory.PODTrajectoryModule(ref_time.m_as("ms"), ref_pos.m_as("m"),
n_modes=10, poly_order=10,
batch_size=batch_size, is_periodic=True)
breathing_module = cmrsim.trajectory.SimpleBreathingMotionModule(contraction_module, breathing_curve, ...)
module_acquisition = cmrsim.bloch.GeneralBlochOperator(time_grid=time,
gradient_waveforms=grad_grid,
rf_waveforms=rf_grid,
adc_events=adc_on_grid, ...)
Run Simulation#
[ ]:
for batch_index, batch in tqdm(input_dataset):
m, r = batch.pop("magnetization"), batch.pop("initial_position")
for tr_index in range(n_tr):
m0 = proton_density_module(initial_position,
start_of_trs[tr_index],
batch_index=batch_index)
batch["M0"] = m0
m, r = module_acquisition(initial_position=r, magnetization=m,
repetition_index=tr_index,
trajectory_module=breathing_module, **batch)