Multiple Coils: Using the ParallelReceiveBlochOperator#
In this notebook, the incorporation of coil-sensitivities into Bloch-simulation is highlighted. To do so, first a 3-dimensional coil-sensitivity map is computed assuming an ideal Birdcage geometry based on the implementation of sigpy (link). The coil-maps can be specified as vtk-mesh or numpy arrays, therefore it is trivial to incorporate the results from proper Biot-Savart simulations using third-party
packages.
The phantom used as simulation input, is a sphere with air-enclosures. The sequence used for simulation is a balanced steady state free precession where $B_0$-inhomogeneities are included as demonstrated in the dedicated example notebook.
The notebooks content is summarized as: 1. Imports 2. Create Coil-Sensitivities 3. Define Sequence (Parameters) 4. Construct Phantom (and illustrate) 5. Instatiate Simulation Operators 6. Reconstruct and plot
Imports#
[ ]:
!pip install cmrseq --index-url https://gitlab.ethz.ch/api/v4/projects/30300/packages/pypi/simple
[1]:
import sys
sys.path.append("../")
sys.path.insert(0, "../../")
sys.path.insert(0, "../../../cmrseq")
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
os.environ['CUDA_VISIBLE_DEVICES'] = "3"
import tensorflow as tf
gpu = tf.config.get_visible_devices("GPU")
if gpu:
tf.config.experimental.set_memory_growth(gpu[0], True)
[2]:
import sys
sys.path.insert(0, "../../")
from copy import deepcopy
import base64
from IPython.display import display, HTML, clear_output
import pyvista
from pint import Quantity
import numpy as np
from tqdm.notebook import tqdm
import matplotlib.pyplot as plt
%matplotlib inline
import cmrsim
import cmrseq
sys.path.append("../")
import local_functions
clear_output()
Construct Coil-Sensitivities#
To incorporate the coil-maps into the operator later on, an instance of the CoilSensitivity class must be created. The class method from_3d_birdcage provides a convenient wrapper for the bird-cage geometry. For more information on arbitrary coil-geometries consult the API-reference.
[3]:
coil_module = cmrsim.analytic.contrast.CoilSensitivity.from_3d_birdcage(map_shape=(100, 100, 150, 8),
map_dimensions=Quantity([[-15, 15], [-15, 15], [-20, 20]], "cm").m_as("m"),
relative_coil_radius=1.,
device="GPU:0")
Define Sequence#
General Parameter Definition#
[4]:
# Define Fourier-Encoding parameters
fov = Quantity([25, 25], "cm")
# matrix_size = (151, 151)
matrix_size = (75, 75)
res = fov / matrix_size
print("Resolution:", res)
n_dummy = 50
center_index = np.array(matrix_size) // 2 + np.mod(matrix_size, 2)
# Define imaging slice parameters
slice_thickness = Quantity(6, "mm")
slice_position = Quantity([0., 0., 0.], "m")
adc_duration = Quantity(2., "ms")
readout_direction = np.array([1., 0., 0.])
slice_normal = np.array([0., 0., 1.])
slice_position_offset = Quantity(0., "cm")
# Define RF-exication parameters (coupled with seeding slice atm)
flip_angle = Quantity(np.pi/4, "rad")
pulse_duration = Quantity(0.6, "ms")
Resolution: [0.3333333333333333 0.3333333333333333] centimeter
Instantiate sequence#
[5]:
# Define MR-system specifications
system_specs = cmrseq.SystemSpec(max_grad=Quantity(50, "mT/m"), max_slew=Quantity(200., "mT/m/ms"),
grad_raster_time=Quantity(0.01, "ms"),
rf_raster_time=Quantity(0.01, "ms"),
adc_raster_time=Quantity(0.01, "ms"),
b0=Quantity(1.5, "T"))
seq_list = cmrseq.seqdefs.sequences.balanced_ssfp(system_specs,
matrix_size,
repetition_time=Quantity(0., "ms"), # Optimizes for shortest
inplane_resolution=res,
slice_thickness=slice_thickness,
adc_duration=adc_duration,
flip_angle=flip_angle,
pulse_duration=pulse_duration,
slice_position_offset=slice_position_offset,
dummy_shots=n_dummy)
CMRSeq Warning : bSSFP Sequence: Repetition time too short to be feasible, set TR to 3.8800000000000003 millisecond
Illustrate sequence#
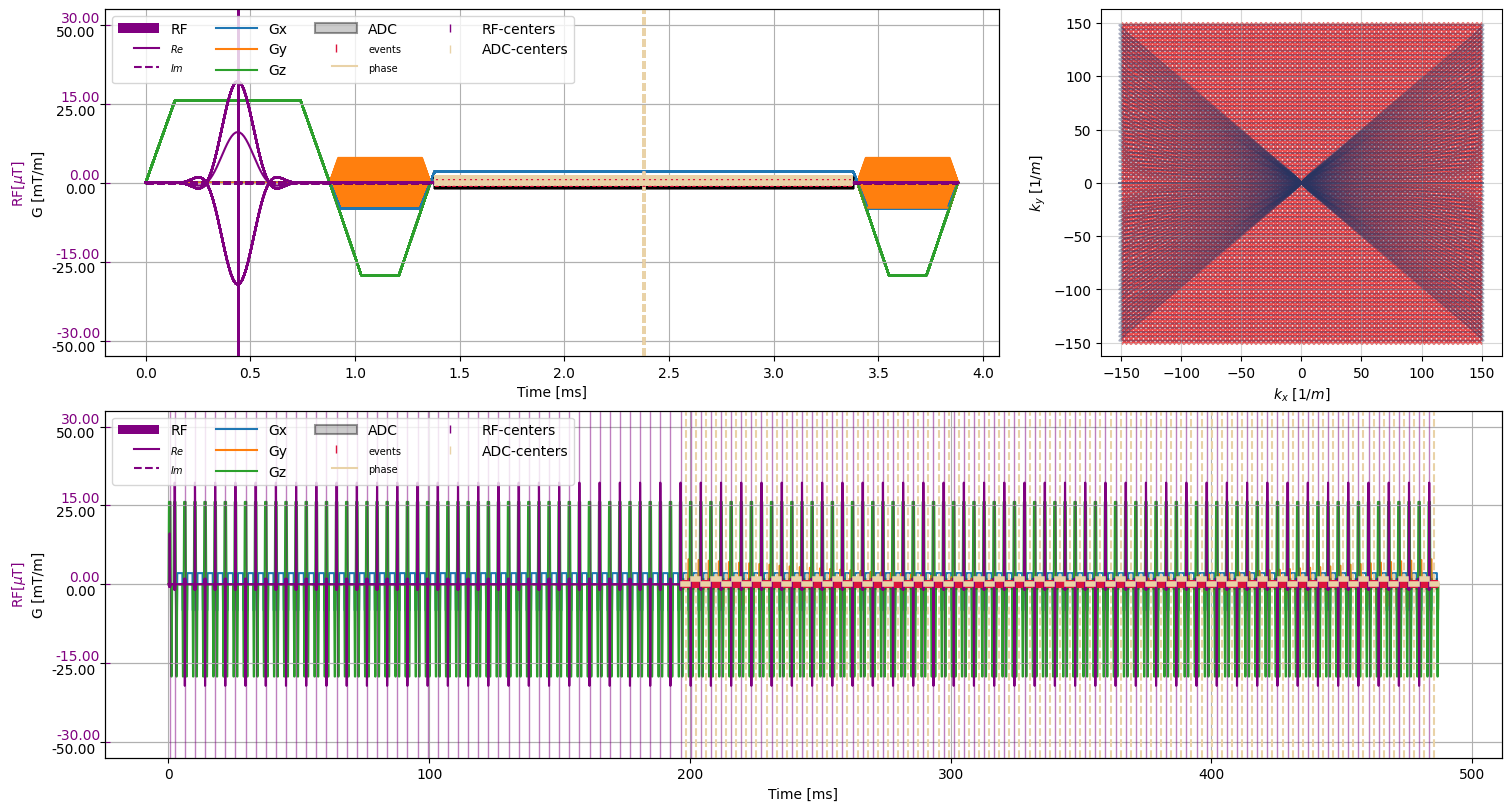
[6]:
plt.close("all")
f = plt.figure(constrained_layout=True, figsize=(15, 8))
axes = f.subplot_mosaic("AAB;CCC")
cmrseq.plotting.plot_sequence(seq_list[0], axes=axes["A"], format_axes=True, add_legend=False)
for s in seq_list[n_dummy:]:
cmrseq.plotting.plot_sequence(s, axes=[f.axes[-1], axes["A"], axes["A"], axes["A"]], format_axes=False)
for s in seq_list[n_dummy:]:
cmrseq.plotting.plot_kspace_2d(s, ax=axes["B"], k_axes=[0, 1])
full_seq = deepcopy(seq_list[0])
full_seq.extend(deepcopy(seq_list[1:]))
cmrseq.plotting.plot_sequence(full_seq, axes=axes["C"])
Extending Sequence: 100%|██████████| 125/125 [00:00<00:00, 213.46it/s]
Construct Phantom#
The geometrical relation of the phantom is defined in the custom function create_spherical_phantom. The RegularGridDataset class is used to compute the off-resonance maps as well as selecting a slice from the phantom to reduces the computational load during simulation (for isochromates permanently outside the excitation profile)
[7]:
phantom_ = local_functions.create_spherical_phantom(dimensions=(61, 61, 61), spacing=(0.004, 0.004, 0.004), big_radius=0.12)
phantom_["in_mesh"] = np.ones(phantom_.points.shape[0])
phantom = cmrsim.datasets.RegularGridDataset.from_unstructured_grid(phantom_, pixel_spacing=Quantity([1, 1, 1], "mm"),
padding=Quantity([5, 5, 5], "cm"))
phantom.mesh["chi"] = np.where(phantom.mesh["in_mesh"], np.ones_like(phantom.mesh["in_mesh"]) * (-9.05),
np.ones_like(phantom.mesh["in_mesh"]) * 0.36) * 1e-6 # susceptibilty in ppm
phantom.compute_offresonance(b0=system_specs.b0, susceptibility_key="chi")
clear_output()
voxels = phantom.select_slice(slice_normal, spacing=Quantity([0.25, 0.25, 2], "mm"),
slice_position=slice_normal*slice_position_offset,
field_of_view=Quantity([*fov.m_as("m"), slice_thickness.m_as("m")*1.5], "m"),
readout_direction=readout_direction, in_mps=False)
voxels = voxels.threshold(0.1, "in_mesh")
CMRSeq Warning : Optimal rotation is not uniquely or poorly defined for the given sets of vectors.
Illustrate the phantom within the coil-maps#
[8]:
# Create a clipped cylindrical mesh to 3D-render the coil-sensitvities
coil_maps = coil_module.coil_maps
coilmaps_magn = {f"mag_coil_{i}": np.abs(coil_maps[..., i]) for i in range(coil_maps.shape[-1])}
coilmaps_phase = {f"phi_coil_{i}": np.angle(coil_maps[..., i]) * 0 for i in range(coil_maps.shape[-1])}
coil_maps = dict(**coilmaps_magn, **coilmaps_phase)
coil_dataset = cmrsim.datasets.RegularGridDataset.from_numpy(data=coil_maps, extent=Quantity([60, 60, 90], "cm"))
cylinder = pyvista.Cylinder(center=(0., 0., 0.), direction=(0., 0., 1.), radius=0.15, height=0.4)
cylinder = coil_dataset.mesh.clip_surface(cylinder.extract_surface())
full_sphere = phantom_.sample(coil_dataset.mesh)
[9]:
import imageio
# Create the images
images = []
for coil_id in range(8):
pyvista.close_all()
pyvista.start_xvfb()
plotter = pyvista.Plotter(off_screen=True, window_size=(300, 300))
plotter.add_mesh(voxels, color="white")
plotter.add_mesh(full_sphere, scalars=f"mag_coil_{coil_id}", cmap="plasma", opacity=0.9)
plotter.add_mesh(cylinder, scalars=f"mag_coil_{coil_id}", cmap="plasma", show_scalar_bar=False, opacity=0.8)
local_functions.add_custom_axes(plotter, scale=0.3)
plotter.add_title(f"Coil #{coil_id}", font_size=8)
images.append(plotter.screenshot("coil_rendering.png"))
ims = np.concatenate(np.concatenate(np.stack(images).reshape(2, 4, 300, 300, 3), axis=1), axis=1)
imageio.v3.imwrite("coil_rendering.png", ims)
b64 = base64.b64encode(open("coil_rendering.png", 'rb').read()).decode('ascii')
display(HTML(f'<img src="data:image/gif;base64,{b64}" />'))
Instantiate Simulation Operators#
The dummy-shots of the sequence should not be run using the ParallelReceiveBlochOperator, because no sampling events are defined. Therefore, the sequnce definition is split at the first non-dummy shot. The first part is used to instantiate a GeneralBlochOperator while the second part is used to instantiate a ParallelReceiveBlochOperator which takes the coil-sensitivities created above as argument.
[10]:
# Split the dummyshots and create single Sequence instance
dummy_sequence = deepcopy(seq_list[0])
dummy_sequence.extend(seq_list[1:n_dummy+1])
# Grid the sequence definitions
time_dummy, rf_grid_dummy, grad_grid_dummy, _ = [np.stack(v) for v in cmrseq.utils.grid_sequence_list([dummy_sequence, ])]
time, rf_grid, grad_grid, adc_on_grid = [np.stack(v) for v in cmrseq.utils.grid_sequence_list(seq_list[n_dummy+1:])]
# Define off-resonsance submodule
offres = cmrsim.bloch.submodules.OffResonance(gamma=system_specs.gamma.m_as("MHz/T"), device="GPU:0")
# Construct BlochOperators and pass offresonance module:
module_dummyshots = cmrsim.bloch.GeneralBlochOperator(name="dummy_shots", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
time_grid=time_dummy[0],
gradient_waveforms=grad_grid_dummy,
rf_waveforms=rf_grid_dummy,
device="GPU:0",
submodules=[offres, ]
)
module_acquisition = cmrsim.bloch.ParallelReceiveBlochOperator(name="acquisition", gamma=system_specs.gamma_rad.m_as("rad/mT/ms"),
coil_module=coil_module,
time_grid=time[0],
gradient_waveforms=grad_grid,
rf_waveforms=rf_grid,
adc_events=adc_on_grid,
device="GPU:0",
submodules=[offres, ]
)
Extending Sequence: 100%|██████████| 50/50 [00:00<00:00, 184.86it/s]
Prepare simulation input#
[15]:
n_particles = voxels.points.shape[0]
voxels["M0"] = np.ones(n_particles, dtype=np.float32)
voxels["T1"] = np.ones(n_particles, dtype=np.float32) * 1000
voxels["T2"] = np.ones(n_particles, dtype=np.float32) * 200
slice_dataset = cmrsim.datasets.RegularGridDataset(voxels)
properties = slice_dataset.get_phantom_def(filter_by="in_mesh", keys=["M0", "T1", "T2", "offres"],
perturb_positions_std=Quantity(0.1, "mm").m_as("m"))
properties["initial_position"] = properties.pop("r_vectors")
properties["off_res"] = properties.pop("offres").reshape(-1, 1)
properties["magnetization"] = cmrsim.utils.particle_properties.norm_magnetization()(n_particles)
input_dataset = cmrsim.datasets.BlochDataset(properties, filter_inputs=True)
print([v["initial_position"].shape for v in input_dataset(batchsize=int(3e6)).take(1)])
[TensorShape([2575744, 3])]
Run simulation#
[16]:
# Instantiate simulator:
print(f"Total time-steps per TR: {time.shape[1]}")
for batch in tqdm(input_dataset(batchsize=int(3e6)).take(1)):
print({k: v.shape for k, v in batch.items()})
m_init = batch.pop("magnetization")
initial_position = batch.pop("initial_position")
m, r = module_dummyshots(initial_position=initial_position, magnetization=m_init, **batch)
for tr_index in tqdm(range(time.shape[0]), desc="Iterating TRs", leave=False):
m, r = module_acquisition(initial_position=r, magnetization=m, repetition_index=tr_index, **batch)
Total time-steps per TR: 439
{'M0': TensorShape([2575744]), 'T1': TensorShape([2575744]), 'T2': TensorShape([2575744]), 'initial_position': TensorShape([2575744, 3]), 'off_res': TensorShape([2575744, 1]), 'magnetization': TensorShape([2575744, 3])}
Reconstruction#
First generate coil-map images by probing the 3D sensitivity maps (again using the functionality provided by the RegularGridDataset class).
[17]:
# Create coil-maps from the original input mesh
sl_mesh = coil_dataset.select_slice(slice_normal, spacing=Quantity([*res.m_as("m"), slice_thickness.m_as("m")], "m"),
slice_position=slice_normal*slice_position_offset,
field_of_view=Quantity([*fov.m_as("m"), slice_thickness.m_as("m")], "m"),
readout_direction=readout_direction, in_mps=True)
coil_sensitivity_images = np.stack([sl_mesh[f"mag_coil_{i}"] + 1j * sl_mesh[f"phi_coil_{i}"] for i in range(8)], axis=-1).reshape(*matrix_size, 8, order="F")
CMRSeq Warning : Optimal rotation is not uniquely or poorly defined for the given sets of vectors.
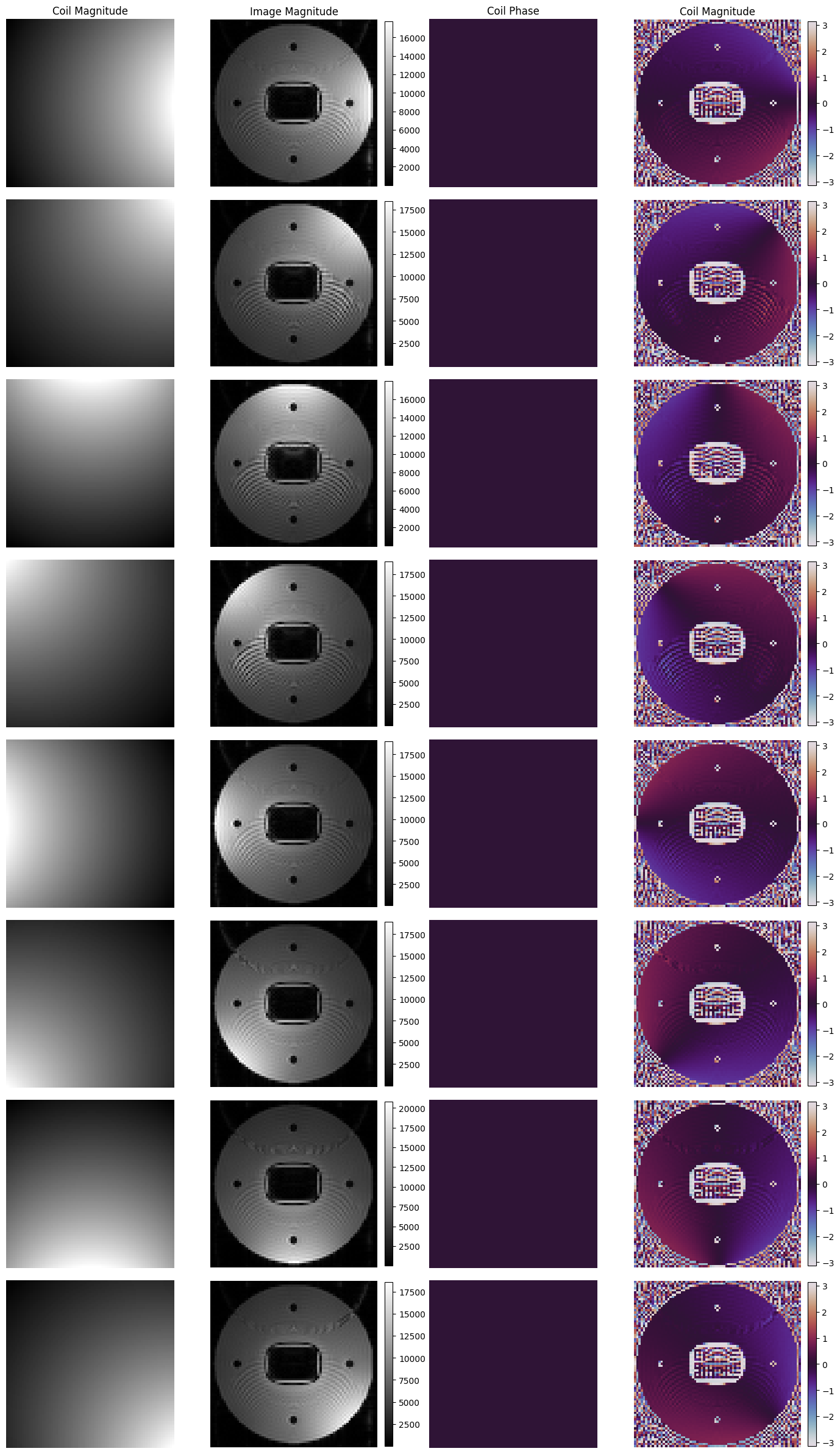
Now reconstruct each coil-channel separately and plot the result side-by-side with the reference sensitivity maps
[18]:
# Show images
fig, axes = plt.subplots(8, 4, figsize=(14, 24))
for i in range(8):
time_signal = tf.stack([v[:, i] for v in module_acquisition.time_signal_acc], axis=0).numpy().reshape(*matrix_size[::-1], order="C")
time_signal += tf.complex(*[tf.random.normal(shape=time_signal.shape, stddev=40) for i in range(2)])
centered_projection = tf.signal.fft(time_signal).numpy()
centered_k_space = tf.signal.fft(centered_projection).numpy()
image = tf.signal.fftshift(tf.signal.ifft2d(tf.roll(tf.signal.ifftshift(centered_k_space, axes=(0, 1)), -1, axis=1)), axes=(0, 1)).numpy() * (0 + 1j)
coil_mag = axes[i, 0].imshow(np.abs(coil_sensitivity_images[..., i]).T, cmap="gray")
abs_plot = axes[i, 1].imshow(np.abs(np.squeeze(image)), cmap="gray")
fig.colorbar(abs_plot, ax=axes[i, 1], fraction=0.045, pad=0.04)
coil_phase = axes[i, 2].imshow(np.angle(coil_sensitivity_images[..., i]).T, cmap="twilight", vmin=-np.pi, vmax=np.pi)
phase_plot = axes[i, 3].imshow(np.angle(np.squeeze(image)), cmap="twilight", vmin=-np.pi, vmax=np.pi)
fig.colorbar(phase_plot, ax=axes[i, 3], fraction=0.045, pad=0.04)
[_.axis("off") for _ in axes[i]]
[_.set_title(t) for _, t in zip(axes[0], ["Coil Magnitude", "Image Magnitude", "Coil Phase", "Coil Magnitude"])]
fig.tight_layout()
[ ]: