Basics of performing Analytic Simulations#
This notebooks aims to show only the basic concepts of analytic simulations inside the cmrsim frame work. The incorporation of motion is shown in the next notebook.
Imports#
[3]:
import sys
sys.path.append("../")
sys.path.insert(0, "../../")
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
os.environ['CUDA_VISIBLE_DEVICES'] = "3"
import tensorflow as tf
gpu = tf.config.get_visible_devices("GPU")
if gpu:
tf.config.experimental.set_memory_growth(gpu[0], True)
[8]:
# 3rd Party dependencies
from pint import Quantity
import pyvista
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# Project library cmrsim
import cmrsim
import local_functions
Define the simulation#
_build method, where the composition of building blocks is performed. As the simulation parameters are defined as tf.Variables their values are easily modifyable at any time, even with graph execution._build member-function must return a triple containing an instance of a subclassed EncodingBase module, a CompositeSignalModel (composing all signal operator building blocks) and an instance of a ReconBase module. If the simulation is meant to return the k-space signal, the third entry of the returned triple can be set to None.The following ExampleSimulation is setup as a single shot (EPI) spin-echo, considering the T2* effects (TE-t, TE+t) with a simple inverse FFT as reconstruction.
[5]:
from typing import Tuple
class ExampleSimulation(cmrsim.analytic.AnalyticSimulation):
def build(self, matrix_size: Tuple[int, int] = (80, 100),
field_of_view: Tuple[float, float] = (0.25, 0.3)):
n_kx, n_ky = matrix_size
# Encoding definition / To enable result comparisons, the noise is set to 0 here
encoding_module = cmrsim.analytic.encoding.EPI(field_of_view=field_of_view, sampling_matrix_size=matrix_size,
read_out_duration=0.4, blip_duration=0.01, k_space_segments=1,
absolute_noise_std=0.)
# Signal module construction
spinecho_module = cmrsim.analytic.contrast.SpinEcho(echo_time=50, repetition_time=10000, expand_repetitions=True)
centered_times = tf.abs(encoding_module.get_sampling_times() - tf.reduce_max(encoding_module.get_sampling_times()) / 2)
t2_star_module = cmrsim.analytic.contrast.t2_star.StaticT2starDecay(centered_times, expand_repetitions=False)
# Forward model composition
forward_model = cmrsim.analytic.CompositeSignalModel(spinecho_module, t2_star_module)
# Reconstruction
recon_module = cmrsim.reconstruction.Cartesian2D(sample_matrix_size=(n_kx, n_ky), padding=None)
return forward_model, encoding_module, recon_module
To run the simulation, a Simulator-object needs to be instantiated.
[6]:
field_of_view = Quantity((0.25, 0.3), "m")
matrix_size = (80, 100)
simulator = ExampleSimulation(name='simulation_example', build_kwargs=dict(matrix_size=matrix_size, field_of_view=field_of_view.m_as("m").tolist()))
CMRSeq DeprecationWarning: /tmp/ipykernel_3220715/888904239.py:10 : Consider using the cmrsim.analytic.encoding.GenericEncoding class incombination with a cmrseq.sequence definition
Now display what the requirements for the digital phantoms are, i.e. which physical properties must each material point have assigned:
[7]:
simulator.forward_model.required_quantities, simulator.get_k_space_shape()
[7]:
(('M0', 'T1', 'T2', 'T2star'),
<tf.Tensor: shape=(2,), dtype=int32, numpy=array([ 1, 8000], dtype=int32)>)
Define a digital phantom#
In the following cell, a 2D object is constructed by loading a semantic mask (labels are integers) from the XCAT phantom.
cmrsim.utils.coordinates is an easy way to calculate them.[9]:
phantom_mesh = local_functions.create_cylinder_phantom()
clear_output()
phantom_mesh
[9]:
| Header | Data Arrays | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
[10]:
pyvista.close_all()
pyvista.start_xvfb()
plotter = pyvista.Plotter(window_size=(1600, 300), shape=(1, 4))
for idx, sc in enumerate(["class", "M0", "T1", "T2"]):
plotter.subplot(0, idx)
plotter.add_mesh(phantom_mesh.copy(), scalars=sc)
local_functions.add_custom_axes(plotter, scale=0.2)
box = local_functions.transformed_box(reference_basis=np.eye(3, 3), slice_normal=np.array([0., 0., 1.]), readout_direction=np.array([1., 0., 0.]),
slice_position=Quantity([0., 0., 0.], "m"), extend=Quantity([*field_of_view.m_as("m"), 0.01], "m"))
plotter.add_mesh(box, color="white", opacity=0.5)
plotter.show(jupyter_backend="static")
CMRSeq Warning : Optimal rotation is not uniquely or poorly defined for the given sets of vectors.
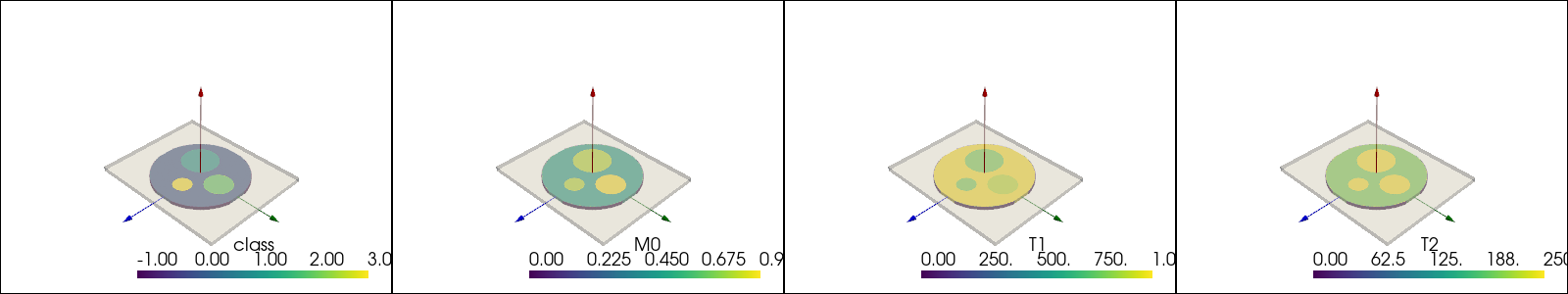
To pass the phantom into the simulator, the a dictionary is used to instantiate a costum tf.data.Dataset implemented in the BaseDataset class. The dictionary contains numpy arrays corresponding to the required quantities listed above.
[11]:
phantom_dict = dict(
M0=phantom_mesh["M0"].astype(np.complex64),
T1=phantom_mesh["T1"].astype(np.float32),
T2=phantom_mesh["T2"].astype(np.float32),
T2star=phantom_mesh["T2star"].astype(np.float32),
r_vectors=phantom_mesh.points.astype(np.float32))
print("Phantom Definition: \t", [f"{k}: {v.shape}" for k, v in phantom_dict.items()])
dataset = cmrsim.datasets.AnalyticDataset(phantom_dict, filter_inputs=True, expand_dimension=True)
Phantom Definition: ['M0: (130204,)', 'T1: (130204,)', 'T2: (130204,)', 'T2star: (130204,)', 'r_vectors: (130204, 3)']
Run the simulation#
To start the simulation, use the __call__ function of the simulator instance {() - operator}.
[12]:
%time simulation_result = simulator(dataset, voxel_batch_size=10000)
simulation_result.shape
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
I0000 00:00:1712952783.398953 3220715 device_compiler.h:186] Compiled cluster using XLA! This line is logged at most once for the lifetime of the process.
Run Simulation: |XXXXXXXXXXXXXXX|127548/127548 |XXXXXXXXXXXXXXX|1/1 CPU times: user 3.46 s, sys: 699 ms, total: 4.16 s
Wall time: 4.31 s
[12]:
TensorShape([1, 100, 80])
[13]:
f, a = plt.subplots(1, 2, figsize=(8, 6))
magnitude_image = np.squeeze(np.abs(simulation_result))
phase_image = np.squeeze(np.angle(simulation_result))
img1 = a[0].imshow(magnitude_image, cmap="gray")
img2 = a[1].imshow(phase_image, cmap="twilight")
[plt.colorbar(im, ax=ax, location="bottom", pad=0.01, shrink=0.9) for im, ax in zip([img1, img2], a)]
[_.axis("off") for _ in a.flatten()]
[_.set_title(t, fontsize=20) for _, t in zip(a, ("Magnitude", "Phase"))]
f.tight_layout()
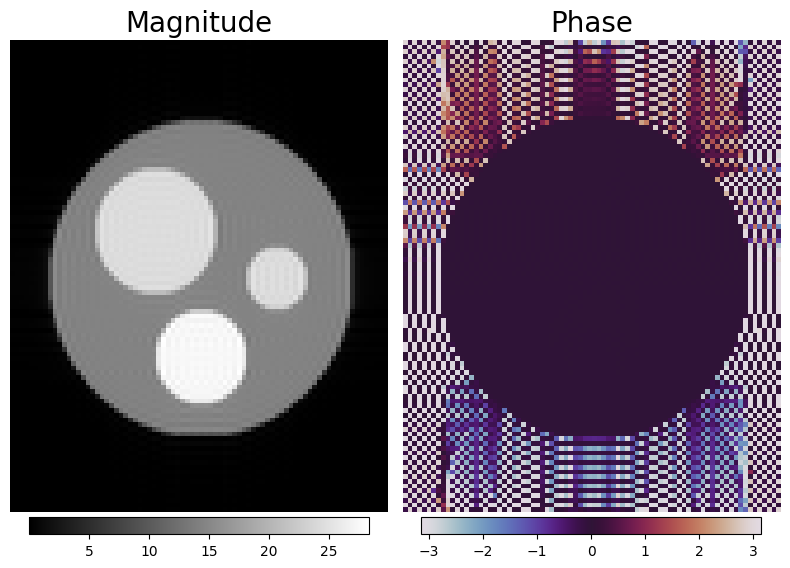
Auxilliary functions#
1. Saving and Loading#
The next cell shows how the simulation with all its configurations can be saved as tf.train.Checkpoint, and afterwards be restored from the checkpoint by calling the classmethod SimulationBase.from_checkpoint(...). To ensure that the parameters are equal, the simulation is redone and the results are compared.
[14]:
import os
# Specify saving path
chkpt = './model_checkpoints/example_checkpoint'
os.makedirs(os.path.dirname(chkpt), exist_ok=True)
# Save simulation to checkpoint and delete old instance
simulator.save(checkpoint_path=chkpt)
del simulator
# construct new intance from checkpoint
new_simulator = ExampleSimulation.from_checkpoint(chkpt)
CMRSeq DeprecationWarning: /tmp/ipykernel_3220715/888904239.py:10 : Consider using the cmrsim.analytic.encoding.GenericEncoding class incombination with a cmrseq.sequence definition
[15]:
%time simulation_load_result = new_simulator(dataset, voxel_batch_size=5000)
print("If this is equal to zero, the simulated results are identical:\n\t Difference of simulated k-space ->",
tf.reduce_sum(simulation_result - simulation_load_result))
Run Simulation: |XXXXXXXXXXXXXXX|127548/127548 |XXXXXXXXXXXXXXX|1/1 CPU times: user 3.05 s, sys: 303 ms, total: 3.35 s
Wall time: 2.99 s
If this is equal to zero, the simulated results are identical:
Difference of simulated k-space -> tf.Tensor((0.073531434-3.5832636e-07j), shape=(), dtype=complex64)
2. Manipulate Parameters#
After instantitation of simulator instances, the variables contained in the submodules can be changed by directly accesing the tf.Variable.assign() method. To show if the results differ after setting different Echo-Times the simulated images are compared again:
[16]:
new_simulator.forward_model.spin_echo.TE.assign([10,]) # in milliseconds
%time n = new_simulator(dataset, voxel_batch_size=5000)
print('Mean absolute difference for different TE:', tf.reduce_mean(tf.abs(simulation_load_result - n)))
new_simulator.forward_model.spin_echo.TE.assign([50,]) # in milliseconds
%time n = new_simulator(dataset, voxel_batch_size=5000)
print('Mean absolute difference for same TE:', tf.reduce_mean(tf.abs(simulation_load_result - n)))
Run Simulation: |XXXXXXXXXXXXXXX|127548/127548 |XXXXXXXXXXXXXXX|1/1 CPU times: user 1.42 s, sys: 139 ms, total: 1.56 s
Wall time: 1.16 s
Mean absolute difference for different TE: tf.Tensor(1.522993, shape=(), dtype=float32)
Run Simulation: |XXXXXXXXXXXXXXX|127548/127548 |XXXXXXXXXXXXXXX|1/1 CPU times: user 1.4 s, sys: 152 ms, total: 1.55 s
Wall time: 1.14 s
Mean absolute difference for same TE: tf.Tensor(0.0, shape=(), dtype=float32)
3. Display summary as JSON#
The SimulationBase class includes a functionality to create a report of the simulation configuration (aka submodule Variables) as dictionary. This dictionary can be used to generate a JSON string
[17]:
import json
from IPython.display import JSON
summary_dict = new_simulator.configuration_summary
JSON(json.dumps(summary_dict))
CMRSeq Warning : JSON expects JSONable dict or list, not JSON strings
[17]:
<IPython.core.display.JSON object>