Analytic Spatially Selective Excitation#
This notebook highlights the analytic contrast module, allowing for spatial selection for simulation. Conceptually this corresponds to a slice selective excitation in the easiest case and an inner volume excitation technique for 3D selection.
Import#
[1]:
import sys
sys.path.append("../")
sys.path.insert(0, "../../")
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
os.environ['CUDA_VISIBLE_DEVICES'] = "3"
import tensorflow as tf
gpu = tf.config.get_visible_devices("GPU")
if gpu:
tf.config.experimental.set_memory_growth(gpu[0], True)
[9]:
# 3rd Party dependencies
from pint import Quantity
import pyvista
import numpy as np
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
from IPython.display import display, HTML, Image, clear_output
%matplotlib inline
# Project library cmrsim
import cmrsim
import cmrseq
import local_functions
Slice selective excitation#
[11]:
uniform_mesh = pyvista.ImageData(spacing=(0.001, 0.001, 0.001), dimensions=(100, 100, 100), origin=(-0.05, -0.05, -0.05))
r_vectors = tf.constant(uniform_mesh.points, dtype=tf.float32)
slice_normal = np.array([[0, 1, 1], [0, 0, 1], [1, 2, 0]], dtype=np.float64)
slice_normal /= np.linalg.norm(slice_normal, keepdims=True, axis=-1)
slice_position = Quantity([[0, 1, 1], [1, 0, 0], [0, 1, 0]], "cm").m_as("m")
slice_thickness = Quantity([2, 4, 0.5], "cm").m_as("m")
slice_mod = cmrsim.analytic.contrast.SliceProfile(expand_repetitions=True, slice_normal=slice_normal, slice_position=slice_position,
slice_thickness=slice_thickness)
r_vectors_excitation = tf.reshape(r_vectors, [-1, 1, 1, 3])
signal_start = tf.ones(r_vectors_excitation.shape[:-1], dtype=tf.complex64)
signal_out = slice_mod(signal_tensor=signal_start, r_vectors_excitation=r_vectors_excitation).numpy()
for i in range(signal_out.shape[1]):
uniform_mesh[f"signal_out{i}"] = np.abs(signal_out)[:, i, 0]
[13]:
pyvista.close_all()
for i in range(signal_out.shape[1]):
pyvista.start_xvfb()
plotter = pyvista.Plotter(off_screen=True, window_size=(400, 600))
plotter.add_mesh(uniform_mesh, scalars=f"signal_out{i}", show_scalar_bar=False)
plotter.add_title(f"{np.around(slice_normal[i], decimals=2)}\n{slice_thickness[i]}", color="gray", font_size=10)
plotter.screenshot(f"slice_select{i}.png");
pyvista.close_all()
import imageio
img = np.concatenate([imageio.v3.imread(f"slice_select{i}.png") for i in range(signal_out.shape[1])], axis=1)
imageio.v3.imwrite("slice_select.png", img)
display(Image("slice_select.png"))

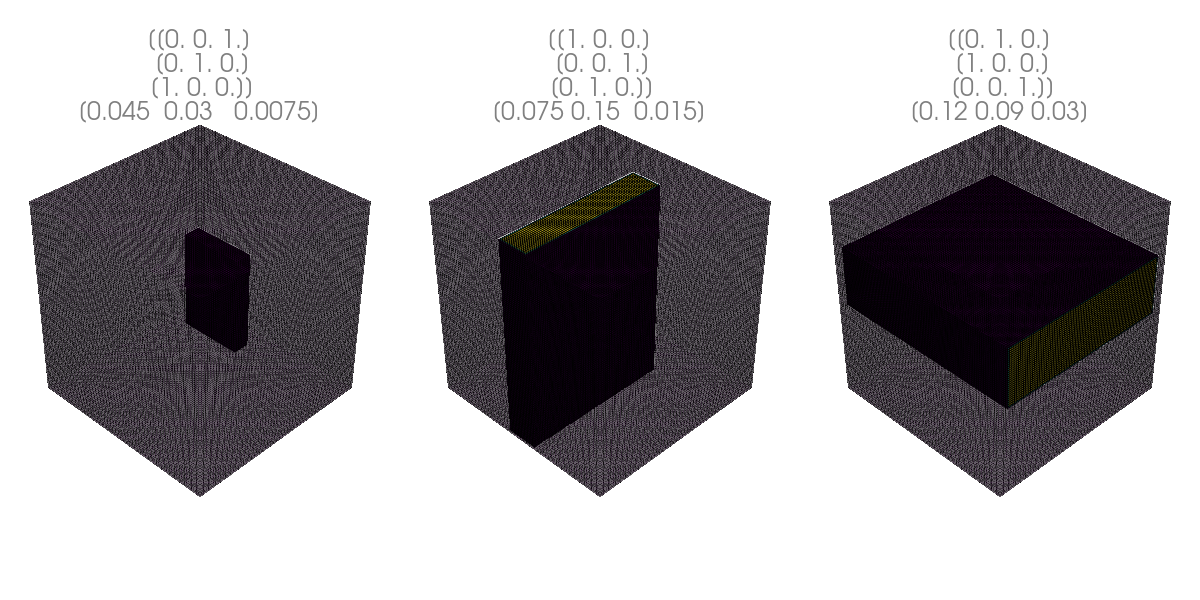
Inner Volume excitation#
[7]:
uniform_mesh = pyvista.ImageData(spacing=(0.001, 0.001, 0.001), dimensions=(100, 100, 100), origin=(-0.05, -0.05, -0.05))
r_vectors = tf.constant(uniform_mesh.points, dtype=tf.float32)
slice_normal = np.eye(3, 3)
readouts = np.roll(np.eye(3, 3), 1, axis=0)
phase_encodes = np.roll(np.eye(3, 3), 2, axis=0)
slice_position = Quantity([[0, 1, 1], [1, 0, 0], [0, 0, 1]], "cm").m_as("m")
spatial_extends = Quantity([[3, 2, 0.5], [5, 10, 1], [8, 6, 2]], "cm").m_as("m") * 1.5
rotation_matrices = tf.stack([readouts, phase_encodes, slice_normal], axis=1)
lolo_mod = cmrsim.analytic.contrast.LocalLookREST(expand_repetitions=True, slice_normal=slice_normal,
readout_direction=readouts, phase_encoding_direction=phase_encodes,
slice_position=slice_position, spatial_extends=spatial_extends)
r_vectors_excitation = tf.reshape(r_vectors, [-1, 1, 1, 3])
signal_start = tf.ones(r_vectors_excitation.shape[:-1], dtype=tf.complex64)
signal_out = lolo_mod(signal_tensor=signal_start, r_vectors_excitation=r_vectors_excitation).numpy()
for i in range(signal_out.shape[1]):
uniform_mesh[f"signal_out{i}"] = np.abs(signal_out)[:, i, 0]
[1000000 1 1] [1000000 3 1]
[10]:
pyvista.close_all()
for i in range(signal_out.shape[1]):
pyvista.start_xvfb()
plotter = pyvista.Plotter(off_screen=True, window_size=(400, 600))
plotter.add_mesh(uniform_mesh, scalars=f"signal_out{i}", show_scalar_bar=False, opacity=0.4, show_edges=True)
plotter.add_mesh(uniform_mesh.threshold(0.1, f"signal_out{i}"), scalars=f"signal_out{i}", show_scalar_bar=False, show_edges=True)
plotter.add_title(f"{np.around(rotation_matrices[i], decimals=2)}\n{spatial_extends[i]}", color="gray", font_size=10)
plotter.screenshot(f"slice_select{i}.png");
pyvista.close_all()
import imageio
img = np.concatenate([imageio.v3.imread(f"slice_select{i}.png") for i in range(signal_out.shape[1])], axis=1)
imageio.v3.imwrite("slice_select.png", img)
display(Image("slice_select.png"))