Introduction Part IV - Parametric Definitions#
Building on the core functionality, cmrseq includes a library of parameterized definitions for commonly occuring groups of building blocks.
This notebooks highlights a few deinitions to demonstrate the principle of usage.
A full list of implemented definitions is given in the API Documentation
Imports#
[1]:
from copy import deepcopy
import numpy as np
from pint import Quantity
import matplotlib.pyplot as plt
import sys
sys.path.insert(0, "../../..")
import cmrseq
Define System Specs#
[2]:
system_specs = cmrseq.SystemSpec(gamma=Quantity(42.575, "MHz/T"),
grad_raster_time=Quantity(5, "us"),
max_grad=Quantity(40, "mT/m"),
max_slew=Quantity(120, "mT/m/ms"),
rf_raster_time=Quantity(5, "us"),
adc_raster_time=Quantity(0.1, "us"))
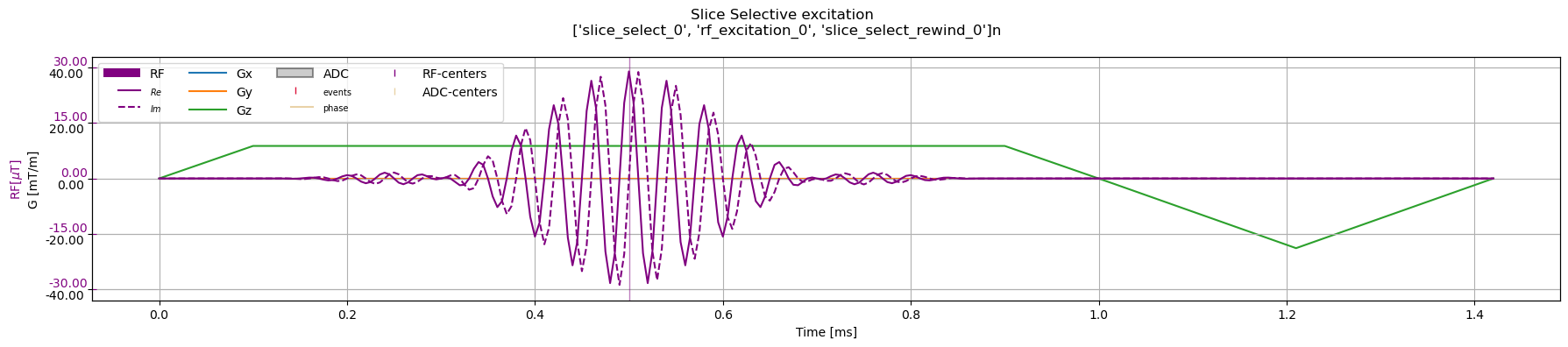
Slice Selective Excitation#
Returns a single sequence object containing gradients and an RF pulse
[3]:
slice_selective_rf = cmrseq.seqdefs.excitation.slice_selective_sinc_pulse(system_specs,
slice_thickness=Quantity(10, "mm"),
flip_angle=Quantity(90, "degree"),
pulse_duration=Quantity(0.8, "ms"),
time_bandwidth_product=4,
delay=Quantity(0, "ms"),
slice_normal=np.array([0., 0., 1.]),
slice_position_offset=Quantity(5, "cm"))
fig, ax = plt.subplots(1, 1, figsize=(18, 4))
cmrseq.plotting.plot_sequence(slice_selective_rf, axes=ax)
fig.suptitle(f"Slice Selective excitation \n {slice_selective_rf.blocks}n")
fig.tight_layout()
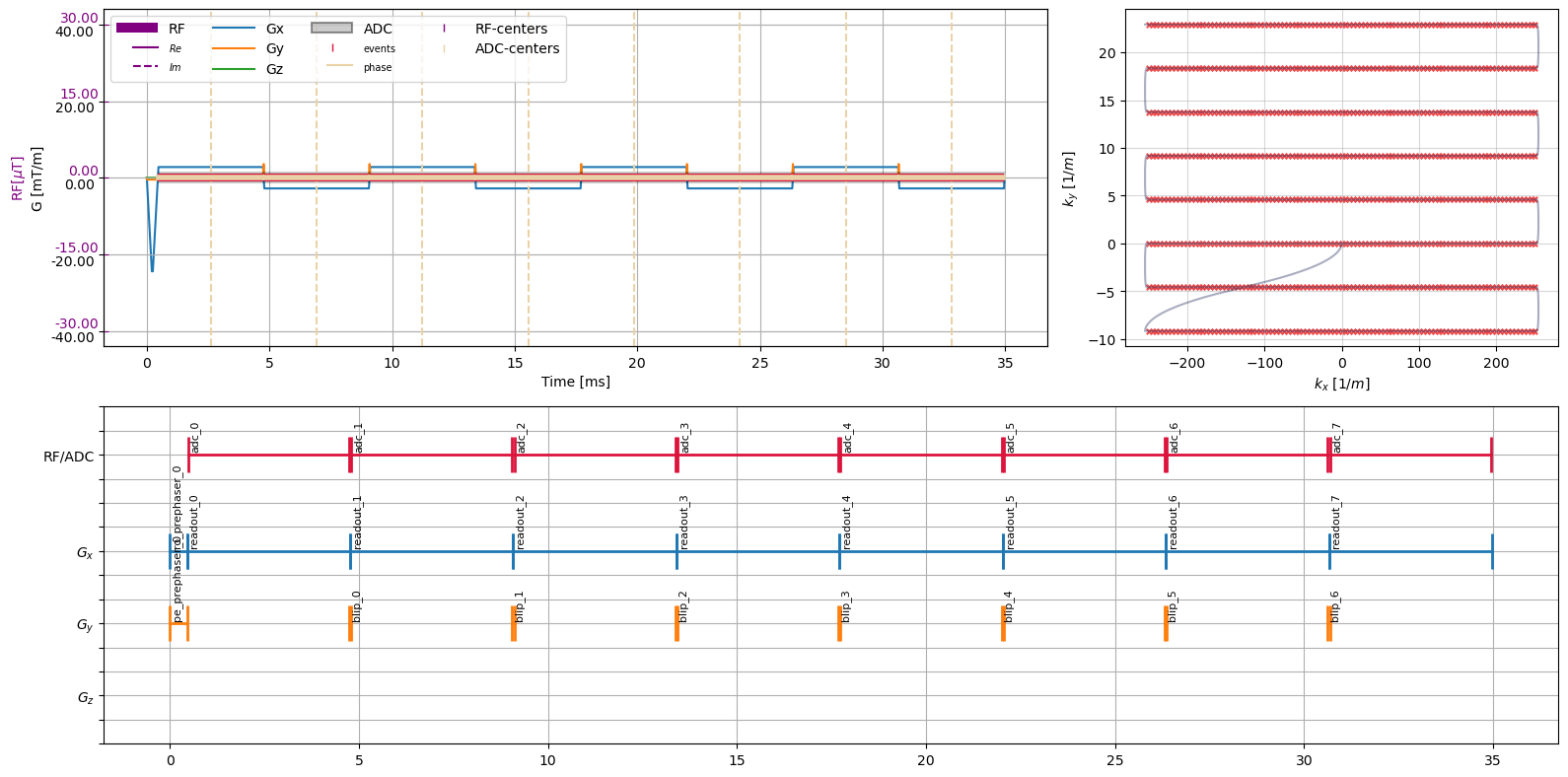
EPI Readout#
[4]:
epi_readout = cmrseq.seqdefs.readout.single_shot_epi(system_specs, field_of_view=Quantity([20, 20], "cm"), matrix_size=np.array([101, 11]),
blip_direction="up", partial_fourier_lines=3, slope_sampling=True, water_fat_shift='maximum',
max_total_duration=Quantity(35, "ms"))
figure = plt.figure(figsize=(16, 8))
axes = figure.subplot_mosaic("AAB;CCC")
axes = np.array([axes[k] for k in "ABC"])
cmrseq.plotting.plot_sequence(seq=epi_readout, axes=axes[0], plot_center_lines=True)
cmrseq.plotting.plot_kspace_2d(seq=epi_readout, k_axes=(0, 1), ax=axes[1])
cmrseq.plotting.plot_block_names(seq=epi_readout, axis=axes[2], fontsize=8)
figure.tight_layout()
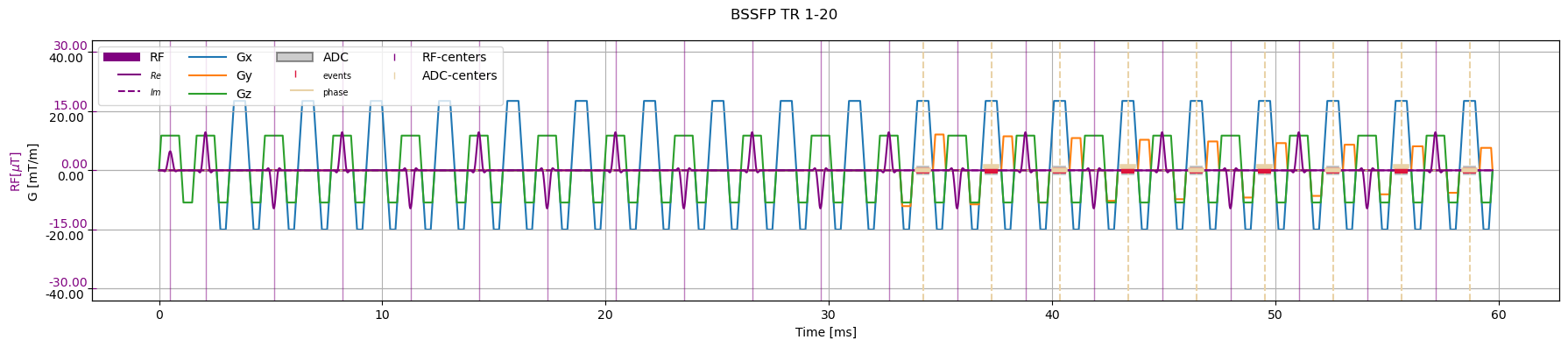
Balanced steady state free precession (BSSFP)#
[5]:
bssfp_list = cmrseq.seqdefs.sequences.balanced_ssfp(system_specs,
inplane_resolution=Quantity([2, 2], "mm"),
matrix_size=np.array([51, 51]),
slice_thickness=Quantity(10, "mm"),
adc_duration=Quantity(0.5, "ms"),
flip_angle=Quantity(30, "degree"),
pulse_duration=Quantity(0.8, "ms"),
repetition_time=Quantity(0, "ms"),
dummy_shots=10)
bssfp = bssfp_list[0]
bssfp.extend(bssfp_list[1:20])
fig, ax = plt.subplots(1, 1, figsize=(18, 4))
cmrseq.plotting.plot_sequence(bssfp, axes=ax)
fig.suptitle("BSSFP TR 1-20")
fig.tight_layout()
/opt/conda/lib/python3.10/site-packages/pint/facets/plain/quantity.py:1345: RuntimeWarning: invalid value encountered in double_scalars
magnitude = magnitude_op(new_self._magnitude, other._magnitude)
/mnt/code/cmrseq/docs/source/getting_started/../../../cmrseq/parametric_definitions/sequences/_ssfp.py:129: UserWarning: Repetition time too short to be feasible, set TR to 3.06 millisecond
warn(f"Repetition time too short to be feasible, set TR to {minimal_tr}")
Extending Sequence: 100%|███████████████████████████████████████████████████████████████████| 19/19 [00:00<00:00, 252.07it/s]