Introduction Part II - Sequence Modification#
This notebooks demonstrates how to modify sequences that have already been instantiated as well as how to combine and extend Sequence objects and building blocks.
Imports#
[1]:
import sys
sys.path.insert(0, "../../..")
import cmrseq
from copy import deepcopy
import numpy as np
from pint import Quantity
import matplotlib.pyplot as plt
%matplotlib inline
Construct Sequence from Part I#
[2]:
system_specs = cmrseq.SystemSpec(gamma=Quantity(42.575, "MHz/T"),
grad_raster_time=Quantity(5, "us"),
max_grad=Quantity(40, "mT/m"),
max_slew=Quantity(120, "mT/m/ms"),
rf_raster_time=Quantity(5, "us"),
adc_raster_time=Quantity(0.1, "us"))
rf_excitation = cmrseq.bausteine.SincRFPulse(system_specs=system_specs,
flip_angle=Quantity(np.pi/2, "rad"),
duration=Quantity(1., "ms"),
time_bandwidth_product=4,
name="fid_excitation")
fov_x = Quantity(30, "cm")
kx_max = 2 / fov_x
prephaser_area = kx_max / system_specs.gamma
ro_prephaser = cmrseq.bausteine.TrapezoidalGradient.from_area(system_specs=system_specs,
area=prephaser_area,
orientation=np.array([-1., 0., 0.]),
name="ro_prephaser")
adc_block = cmrseq.bausteine.SymmetricADC(system_specs=system_specs,
num_samples=50,
duration=Quantity(0.5, "ms"))
ro_gradient = cmrseq.bausteine.TrapezoidalGradient.from_fdur_area(system_specs=system_specs,
orientation=np.array([1., 0., 0.]),
flat_duration=adc_block.duration,
area=prephaser_area*2,
name="ro_gradient")
ro_prephaser.shift(rf_excitation.tmax)
ro_gradient.shift(ro_prephaser.tmax)
adc_block.shift(ro_gradient.tmin + ro_gradient.rise_time)
sequence_obj = cmrseq.Sequence(building_blocks=[rf_excitation, ro_prephaser, ro_gradient, adc_block], system_specs=system_specs)
Modify an existing Sequence#
The building blocks contained inside a sequence object are mutuable, that means modifications of a single block will affect the sequence. All blocks references are stored in a look-up table with unique names, which can be queried via the Sequence.blocks property. Sequences and SequenceBaseBlocks implement the deepcopy operation, therefore if modifications shall not affect the original sequence a copy must be made.
Furthermore, the sequence object implements some properties as (e.g. duration) that queries and processes queries of all contained building blocks.
The following section demontrates a (not-complete) collection of sequence modifications.
Query blocks from sequence#
The are multiple ways to index / query blocks from a sequence object, which can be subdivided into
Query by block names
Index according to temporal ordering
To get information about the contained blocks do the following:
[3]:
print("Blocks contained in sequence: ", sequence_obj.blocks)
print("Start time of sequence", sequence_obj.start_time)
print("End time of sequence", sequence_obj.end_time)
Blocks contained in sequence: ['fid_excitation_0', 'ro_prephaser_0', 'ro_gradient_0', 'adc_0']
Start time of sequence 0.0 millisecond
End time of sequence 1.6 millisecond
1. By names#
[4]:
rf_block = sequence_obj["fid_excitation_0"]
print("By exact name:\n\t", id(rf_block), rf_block.name)
blocks = sequence_obj.get_block(partial_string_match=["fid_", "ro_"])
print("Partial String match:\n\t", "\n\t ".join([f"{id(b)} {b.name}" for b in blocks]))
blocks = sequence_obj.get_block(regular_expression=["ro_.*"])
print("Regular expression match:\n\t", "\n\t ".join([f"{id(b)} {b.name}" for b in blocks]))
By exact name:
140487187816192 fid_excitation
Partial String match:
140487187816192 fid_excitation
140487188021312 ro_prephaser
140487188023568 ro_gradient
Regular expression match:
140487188021312 ro_prephaser
140487188023568 ro_gradient
2. Assume temporal ordering#
[5]:
first_block = sequence_obj[0]
print("Index by one integer:\n\t", id(first_block), first_block.name)
slice_blocks = sequence_obj[0:3]
print("Slice indexing:\n\t", "\n\t ".join([f"{id(b)} {b.name}" for b in slice_blocks]))
arbitrary_blocks = sequence_obj[(0, 2)]
print("Fancy indexing:\n\t", "\n\t ".join([f"{id(b)} {b.name}" for b in arbitrary_blocks]))
all_block_names = [f"{id(b)} {b.name}" for b in sequence_obj]
print("Iterate over Sequence:\n\t", "\n\t ".join(all_block_names))
all_block_names_items = [f"{id(b)} {b.name} {unique_name}" for unique_name, b in sequence_obj.items()]
print("Iterate over Sequence Items:\n\t", "\n\t ".join(all_block_names_items))
Index by one integer:
140487187816192 fid_excitation
Slice indexing:
140487187816192 fid_excitation
140487188021312 ro_prephaser
140487188023568 ro_gradient
Fancy indexing:
140487187816192 fid_excitation
140487188023568 ro_gradient
Iterate over Sequence:
140487187816192 fid_excitation
140487188021312 ro_prephaser
140487188023568 ro_gradient
140487187815040 adc
Iterate over Sequence Items:
140487187816192 fid_excitation fid_excitation_0
140487188021312 ro_prephaser ro_prephaser_0
140487188023568 ro_gradient ro_gradient_0
140487187815040 adc adc_0
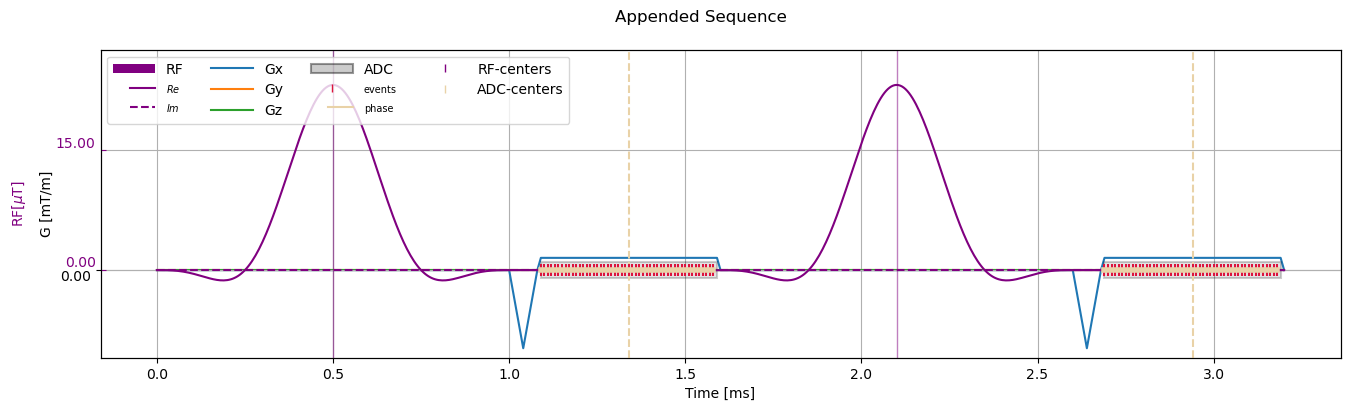
Append Sequence#
Note Appending works with Sequence as well as SequenceBaseBlock objects
The sequence object follows the Python list semantics of append and extend, meaning that single elements are added to the end of the sequence with append and a sorted iterable of sequences can be added more efficiently with extend.
[6]:
repeated_sequence = sequence_obj.copy()
repeated_sequence.append(repeated_sequence, copy=True)
# Visualize
fig, ax = plt.subplots(1, 1, figsize=(16, 4))
cmrseq.plotting.plot_sequence(repeated_sequence, axes=ax)
ax.set_ylim([-4.3889, 11.041])
fig.axes[-1].set_ylim([-10.9721, 27.6025])
_ = fig.suptitle("Appended Sequence")
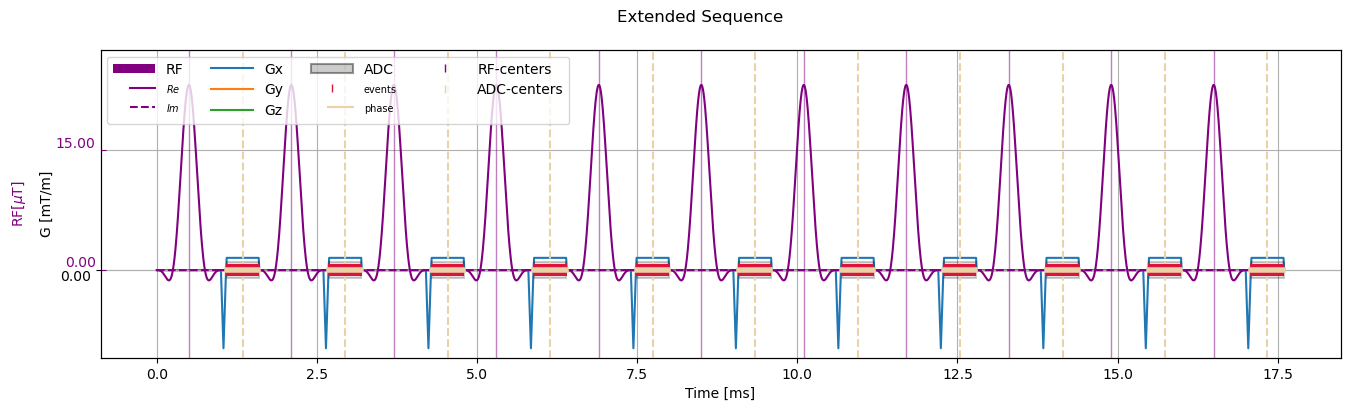
Now add multiple repetitions:
[7]:
repeated_sequence = sequence_obj.copy()
added_repetitions = [sequence_obj.copy() for i in range(10)]
repeated_sequence.extend(added_repetitions, copy=False)
# Visualize
fig, ax = plt.subplots(1, 1, figsize=(16, 4))
cmrseq.plotting.plot_sequence(repeated_sequence, axes=ax)
ax.set_ylim([-4.3889, 11.041])
fig.axes[-1].set_ylim([-10.9721, 27.6025])
_ = fig.suptitle("Extended Sequence")
Extending Sequence: 100%|███████████████████████████████████████████████████████████████████| 10/10 [00:00<00:00, 639.27it/s]
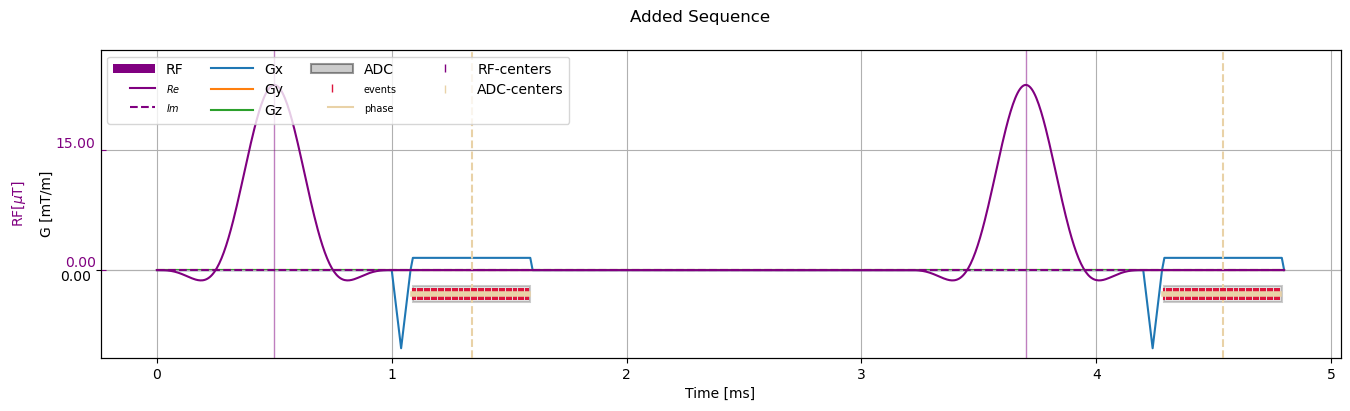
Add Sequences#
If the sequences are alredy correctly shifted corresponding to the same reference point, Sequence objects can be combined by adding them. The regular “+” operator return a new Sequence object where the blocks contained in the summands are deepcopied. The Sequence class also implements the inplace addition “+=”, in which case the sequences are not copied.
[8]:
added_sequence = sequence_obj.copy()
addition = sequence_obj.copy()
addition.shift_in_time(2*added_sequence.duration)
copied_addition = added_sequence + addition
print("Same object after addition?:", copied_addition is added_sequence)
dummy_name = added_sequence
dummy_name += addition
print("Same object after inplace addition?", dummy_name is added_sequence)
# Visualize
fig, ax = plt.subplots(1, 1, figsize=(16, 4))
cmrseq.plotting.plot_sequence(added_sequence, axes=ax, adc_yoffset=-3)
ax.set_ylim([-4.3889, 11.041])
fig.axes[-1].set_ylim([-10.9721, 27.6025])
_ = fig.suptitle("Added Sequence")
Same object after addition?: False
Same object after inplace addition? True
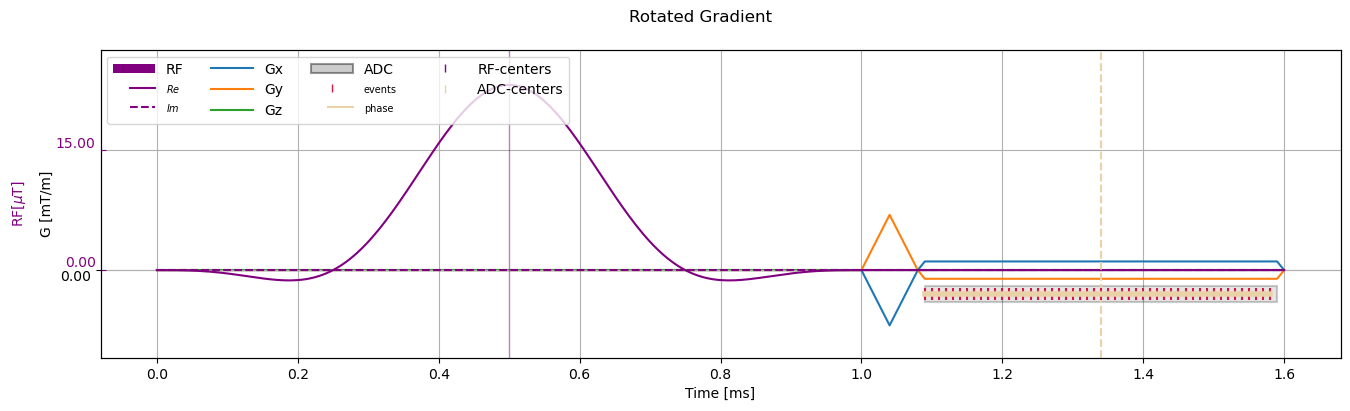
Rotate gradients#
Rotate the gradients from (1., 0., 0.) orientation to (2., 1., 0) / sqrt(5)
[9]:
rotated_sequence = sequence_obj.copy()
rotation_matrix = cmrseq.utils.get_rotation_matrix(slice_normal=np.array([0., 0., 1.]),
readout_direction=np.array([1., 1., 0.]))
rotated_sequence.rotate_gradients(rotation_matrix.T)
# Visualize
fig, ax = plt.subplots(1, 1, figsize=(16, 4))
cmrseq.plotting.plot_sequence(rotated_sequence, axes=ax, adc_yoffset=-3)
ax.set_ylim([-4.3889, 11.041])
fig.axes[-1].set_ylim([-10.9721, 27.6025])
_ = fig.suptitle("Rotated Gradient")
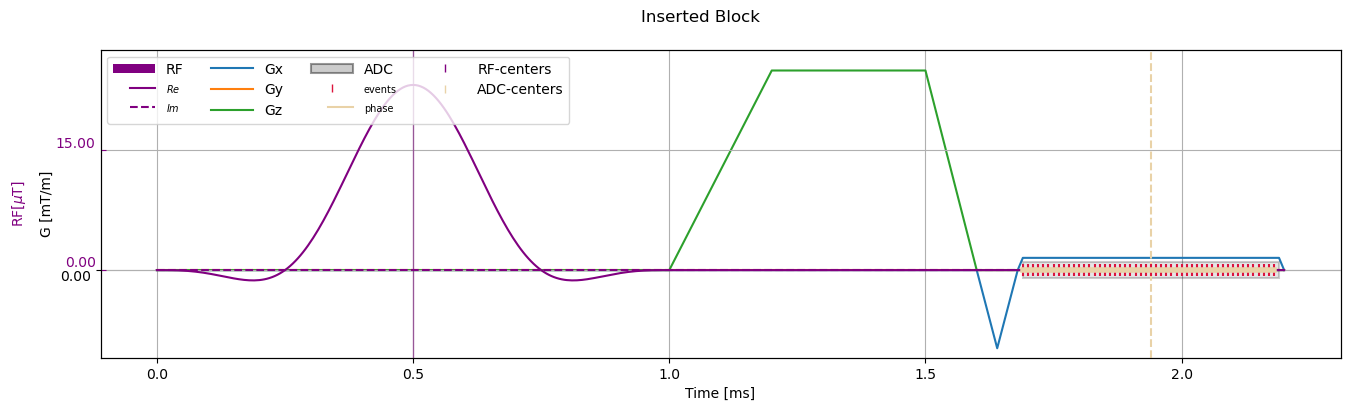
Insert Block#
[10]:
inserted_sequence = sequence_obj.copy()
rf_block = inserted_sequence["fid_excitation_0"]
insert_block = cmrseq.bausteine.TrapezoidalGradient(system_specs, amplitude=Quantity(10, "mT/m"), orientation=np.array([0., 0., 1.]),
rise_time=Quantity(0.2, "ms"), flat_duration=Quantity(0.3, "ms"),
fall_time=Quantity(0.1, "ms"), delay=rf_block.tmax)
for block in inserted_sequence[1:]:
block.shift(insert_block.duration)
inserted_sequence.add_block(insert_block)
# Visualize
fig, ax = plt.subplots(1, 1, figsize=(16, 4))
cmrseq.plotting.plot_sequence(inserted_sequence, axes=ax)
ax.set_ylim([-4.3889, 11.041])
fig.axes[-1].set_ylim([-10.9721, 27.6025])
_ = fig.suptitle("Inserted Block")
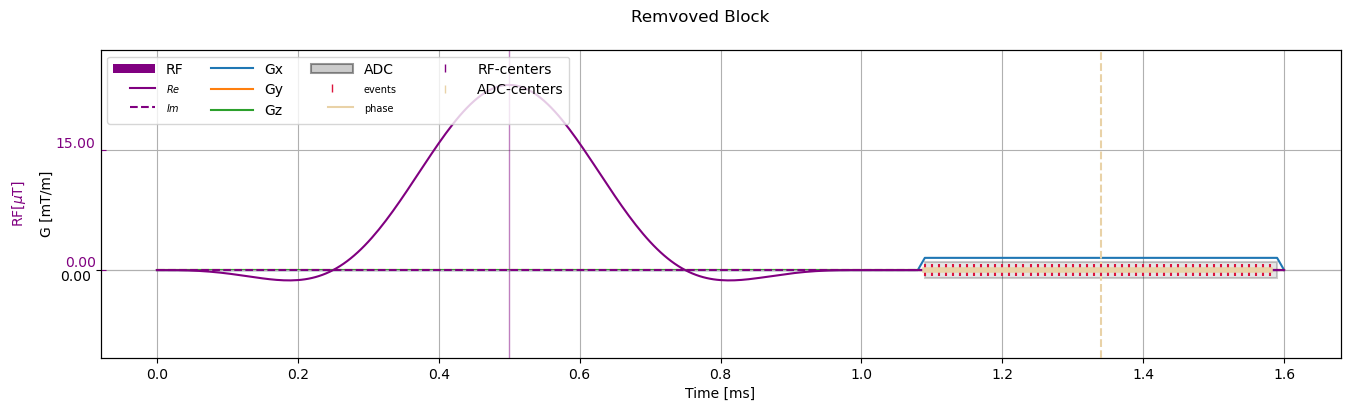
Remove blocks#
[11]:
removed_sequence = sequence_obj.copy()
removed_sequence.remove_block("ro_prephaser_0")
# Visualize
fig, ax = plt.subplots(1, 1, figsize=(16, 4))
cmrseq.plotting.plot_sequence(removed_sequence, axes=ax)
ax.set_ylim([-4.3889, 11.041])
fig.axes[-1].set_ylim([-10.9721, 27.6025])
_ = fig.suptitle("Remvoved Block")