Introduction Part V - Importing / Exporting Sequences#
This notebook shows how to import and export sequence definitions from different file formats.
Currently following file formats are supported:
Pluseq (I/O)
JSON (I/O cmrseq specific)
Phillips GVE (I)
General Imports#
[1]:
import sys
sys.path.insert(0, "../../../")
import os
from copy import deepcopy
import cmrseq
from pint import Quantity
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
PulseSeq#
Getting the sequences to be executed onto the scanner is a key feature for translating simulation experiments to physical measurements. Cmrseq does not implement this feature, but the Pulseq implementation does. Therefore, exporting the defined sequence to the format defined by the Pulseq project offers an indirect way to achieve this functionality. Using the easy way of modifying sequences that cmrseq offers, for pulseq definitions conversion from pulseq to cmrseq Sequence object is implemented.
First Load a sequence definitions stored as test resources: 1. Instantiate a PulSeqFile using the file path 2. Convert to List of cmrseq sequences using to_cmrseq
[2]:
from cmrseq.io import PulseSeqFile
pfile = PulseSeqFile(os.path.abspath(f"../../../tests/resources/trufi.seq"))
sequence_list = pfile.to_cmrseq(gyromagentic_ratio=Quantity(42, "MHz/T"),
max_grad=Quantity(40, "mT/m"),
max_slew=Quantity(200, "mT/m/ms"))
seq1 = deepcopy(sequence_list[0])
seq1.extend(sequence_list[1:], copy=False)
f, a = plt.subplots(1, 1)
cmrseq.plotting.plot_sequence(seq1, axes=a)
plt.show()
0%| | 0/516 [00:00<?, ?it/s]/mnt/code/cmrseq/docs/source/getting_started/../../../cmrseq/core/bausteine/_rf.py:125: UserWarning: When calling snap_to_raster the waveform points are simply rounded to their nearestneighbour if the difference is below the relative tolerance. Therefore this in not guaranteed to be precise anymore
warn("When calling snap_to_raster the waveform points are simply rounded to their nearest"
/mnt/code/cmrseq/docs/source/getting_started/../../../cmrseq/core/bausteine/_gradients.py:159: UserWarning: When calling snap_to_raster the waveform points are simply rounded to their nearestneighbour if the difference is below the relative tolerance. Therefore this in not guaranteed to be precise anymore
warn("When calling snap_to_raster the waveform points are simply rounded to their nearest"
100%|██████████████████████████████████████████████████████████████████████████████████████| 516/516 [00:07<00:00, 67.55it/s]
Extending Sequence: 100%|█████████████████████████████████████████████████████████████████| 515/515 [00:00<00:00, 641.62it/s]
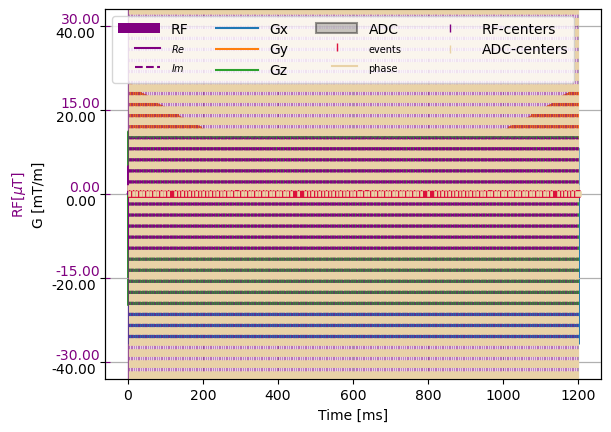
Writing sequences is as easy as loading: 1. Define sequence 2. Instantiate Pulseq file from sequence 3. Call write method
[3]:
from cmrseq.io import PulseSeqFile
system_specs = cmrseq.SystemSpec(max_grad=Quantity(40, "mT/m"), max_slew=Quantity(200., "mT/m/ms"),
grad_raster_time=Quantity(0.01, "ms"), rf_raster_time=Quantity(0.01, "ms"))
fov = Quantity([202, 22], "mm")
matrix_size = np.array((101, 11))
res = fov / matrix_size
adc_duration = Quantity(2., "ms")
pulse_duration = Quantity(1.5, "ms")
slice_thickness = Quantity(2, "cm")
flip_angle = Quantity(12, "degree").to("rad")
print("Resolution:", res)
n_dummy = 0
sequence_list = cmrseq.seqdefs.sequences.flash(system_specs, slice_thickness=slice_thickness,
flip_angle=flip_angle, pulse_duration=pulse_duration,
time_bandwidth_product=6, matrix_size=matrix_size,
inplane_resolution=res, adc_duration=adc_duration,
echo_time=Quantity(2., "ms"), repetition_time=Quantity(10., "ms"),
dummy_shots=n_dummy)
seq1 = sequence_list[0].copy()
seq1.extend(sequence_list[1:], copy=False)
pfile = PulseSeqFile(sequence=seq1)
pfile.write("flash.seq")
/opt/conda/lib/python3.10/site-packages/pint/facets/plain/quantity.py:1345: RuntimeWarning: invalid value encountered in double_scalars
magnitude = magnitude_op(new_self._magnitude, other._magnitude)
Resolution: [2.0 2.0] millimeter
/mnt/code/cmrseq/docs/source/getting_started/../../../cmrseq/parametric_definitions/sequences/_gradient_echo.py:113: UserWarning: Echo time too short to be feasible, set TE to 2.2399999999999998 millisecond
warn(f"Echo time too short to be feasible, set TE to {minimal_te}")
Extending Sequence: 100%|███████████████████████████████████████████████████████████████████| 10/10 [00:00<00:00, 323.32it/s]
JSON#
The json export format is meant to be used to share and store sequence definitions within the cmrseq ecosystem. Saving and loading is done as follows:
[4]:
from IPython.display import JSON
system_specs = cmrseq.SystemSpec(max_grad=Quantity(40, "mT/m"), max_slew=Quantity(200., "mT/m/ms"),
grad_raster_time=Quantity(0.01, "ms"), rf_raster_time=Quantity(0.01, "ms"))
fov = Quantity([202, 22], "mm")
matrix_size = np.array((101, 11))
res = fov / matrix_size
adc_duration = Quantity(2., "ms")
pulse_duration = Quantity(1.5, "ms")
slice_thickness = Quantity(2, "cm")
flip_angle = Quantity(12, "degree").to("rad")
print("Resolution:", res)
n_dummy = 0
sequence_list = cmrseq.seqdefs.sequences.flash(system_specs, slice_thickness=slice_thickness,
flip_angle=flip_angle, pulse_duration=pulse_duration,
time_bandwidth_product=6, matrix_size=matrix_size,
inplane_resolution=res, adc_duration=adc_duration,
echo_time=Quantity(2., "ms"), repetition_time=Quantity(10., "ms"),
dummy_shots=n_dummy)
seq1 = sequence_list[0].copy()
seq1.extend(sequence_list[1:], copy=False)
# Save
cmrseq.io._json.sequence_to_json(seq1, "flash")
# Load
seq = cmrseq.io._json.sequence_from_json("flash.json")
# Display JSON file:
import json
with open("flash.json", "r") as fi:
json_string = json.load(fi)
display(JSON(json_string, expanded=False))
Resolution: [2.0 2.0] millimeter
Extending Sequence: 100%|███████████████████████████████████████████████████████████████████| 10/10 [00:00<00:00, 371.57it/s]
<IPython.core.display.JSON object>
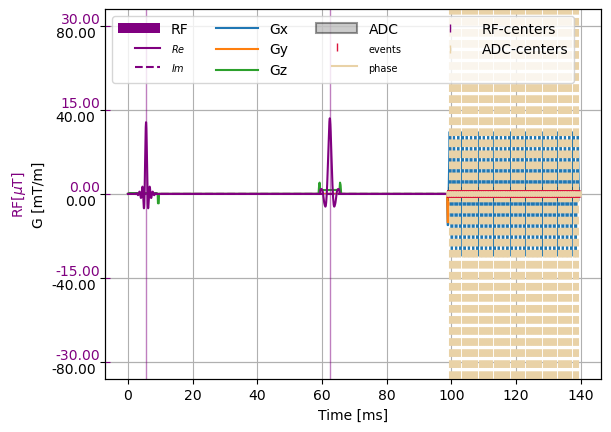
Philips GVE - MPF files#
For the phillips sequence definition, at this moment only loading is supported
[5]:
system_specs = cmrseq.SystemSpec(max_grad=Quantity(80, "mT/m"),
max_slew=Quantity(200., "mT/m/ms"),
grad_raster_time=Quantity(0.001, "ms"),
rf_raster_time=Quantity(0.001, "ms"))
gve_converter = cmrseq.io.GveCmrConvert(system_specs, "../../../tests/resources/diffusion_m012.gve")
Reading sequence objects ...: 100%|████████████████████████████████████████████████████████| 631/631 [00:34<00:00, 18.41it/s]
[6]:
sequence_list = gve_converter(["base_20", "xbase_20"])
imaging_seq = sequence_list[0] + sequence_list[1]
imaging_seq.shift_in_time(-imaging_seq[0].tmin)
Converting to sequence: 100%|██████████████████████████████████████████████████████████████████| 2/2 [00:00<00:00, 3.14it/s]
[7]:
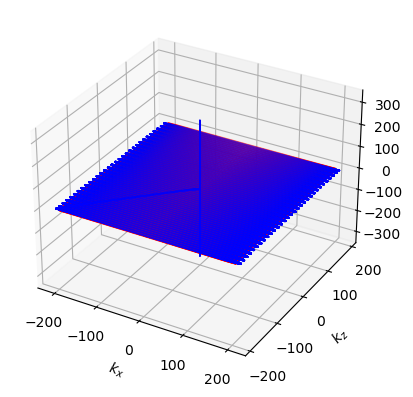
f, a = plt.subplots(1, 1)
cmrseq.plotting.plot_sequence(imaging_seq, axes=a)
cmrseq.plotting.plot_kspace_3d(imaging_seq, plot_raster_trajectory=True)
[7]:
<Axes3D: xlabel='$k_x$', ylabel='$k_z$'>
[ ]: