Introduction I - Basic Functionality#
Import necessary python packages and cmrsim modules#
[1]:
import sys
sys.path.append("../../tests/")
import tensorflow as tf
physical_devices = tf.config.list_physical_devices('GPU')
if physical_devices:
print("Available GPUS: \n\t", "\n\t".join([str(_) for _ in physical_devices]))
tf.config.experimental.set_visible_devices(physical_devices[0], 'GPU')
tf.config.experimental.set_memory_growth(physical_devices[0], True)
from collections import OrderedDict
import numpy as np
import math
import matplotlib.pyplot as plt
%matplotlib inline
import cmrsim
import phantoms
Available GPUS:
PhysicalDevice(name='/physical_device:GPU:0', device_type='GPU')
Define the simulation#
_build method, where the composition of building blocks is performed. As the simulation parameters are defined as tf.Variables their values are easily modifyable at any time, even with graph execution._build member-function must return a triple containing an instance of a subclassed EncodingBase module, a CompositeSignalModel (composing all signal operator building blocks) and an instance of a ReconBase module. If the simulation is meant to return the k-space signal, the third entry of the returned triple can be set to None.The following ExampleSimulation is setup as a single shot (EPI) spin-echo, considering the T2* effects (TE-t, TE+t) with a simple inverse FFT as reconstruction.
[2]:
from typing import Tuple
class ExampleSimulation(cmrsim.analytic.AnalyticSimulation):
def build(self, matrix_size: Tuple[int, int] = (80, 100),
field_of_view: Tuple[float, float] = (0.25, 0.3)):
n_kx, n_ky = matrix_size
# Encoding definition / To enable result comparisons, the noise is set to 0 here
encoding_module = cmrsim.analytic.encoding.EPI(field_of_view=field_of_view, sampling_matrix_size=matrix_size,
read_out_duration=0.4, blip_duration=0.01, k_space_segments=n_ky,
absolute_noise_std=0.)
# Signal module construction
spinecho_module = cmrsim.analytic.contrast.SpinEcho(echo_time=50, repetition_time=10000)
centered_times = tf.abs(encoding_module.get_sampling_times() - tf.reduce_max(encoding_module.get_sampling_times()) / 2)
t2_star_module = cmrsim.analytic.contrast.t2_star.StaticT2starDecay(centered_times)
# Forward model composition
forward_model = cmrsim.analytic.CompositeSignalModel(spinecho_module, t2_star_module)
# Reconstruction
recon_module = cmrsim.reconstruction.Cartesian2D(sample_matrix_size=(n_kx, n_ky), padding=None)
return forward_model, encoding_module, recon_module
To run the simulation, a Simulator-object needs to be instantiated.
[3]:
simulator = ExampleSimulation(name='simulation_example', build_kwargs=dict(matrix_size=(80, 100), field_of_view=(0.25, 0.3)))
Now display what the requirements for the digital phantoms are, i.e. which physical properties must each material point have assigned:
[4]:
simulator.forward_model.required_quantities
[4]:
('M0', 'T1', 'T2', 'T2star')
Define a digital phantom#
In the following cell, a 2D object is constructed by loading a semantic mask (labels are integers) from the XCAT phantom.
cmrsim.utils.coordinates is an easy way to calculate them.[5]:
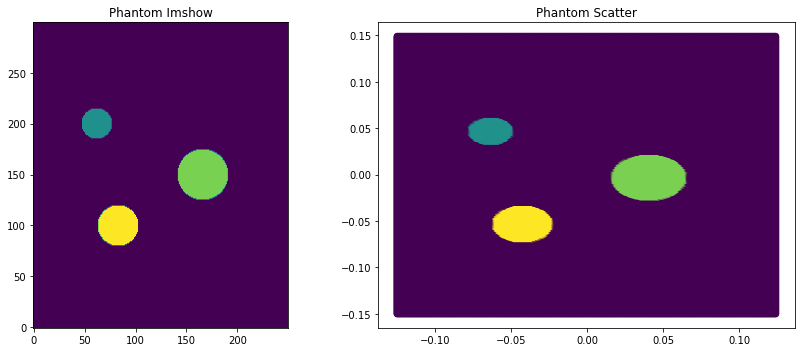
phantom_dict, phantom_reference_map, coordinates = phantoms.define_cylinder_phantom((0.25, 0.3), (250, 300), background_value=0)
f, a = plt.subplots(1, 2, figsize=(12, 5))
a[0].imshow(phantom_reference_map, origin="lower")
a[1].scatter(coordinates[..., 0], coordinates[..., 1], c=np.abs(phantom_dict["M0"].reshape(-1)))
[_.set_title(t) for _, t in zip(a, ["Phantom Imshow", "Phantom Scatter"])]
f.tight_layout()
BaseDataset class.[6]:
for k, v in phantom_dict.items():
if k != "r_vectors":
phantom_dict[k] = v.reshape(-1)
print("Phantom Definition: \t" , [f"{k}:{v.shape}" for k, v in phantom_dict.items()])
dataset = cmrsim.datasets.AnalyticDataset(phantom_dict, filter_inputs=True, expand_dimension=True)
Phantom Definition: ['M0:(75000,)', 'r_vectors:(75000, 3)', 'T1:(75000,)', 'T2:(75000,)', 'T2star:(75000,)']
Run the simulation#
__call__ function of the simulator instance {() - operator}.If the phatom gets larger (more material points), the time for building the Graph will get negligible, thus executing the simulation in eager mode is not advisable.
[7]:
%time simulation_result = simulator(dataset, voxel_batch_size=20000, execute_eager=True)
%time simulation_result = simulator(dataset, voxel_batch_size=20000, execute_eager=False)
Run Simulation: |XXXXXXXXXXXXXXX|3883/3883 |XXXXXXXXXXXXXXX|100/100 CPU times: total: 4.25 s
Wall time: 3.63 s
Run Simulation: |XXXXXXXXXXXXXXX|3883/3883 |XXXXXXXXXXXXXXX|100/100 CPU times: total: 2.92 s
Wall time: 2.48 s
[8]:
import ipywidgets as widgets
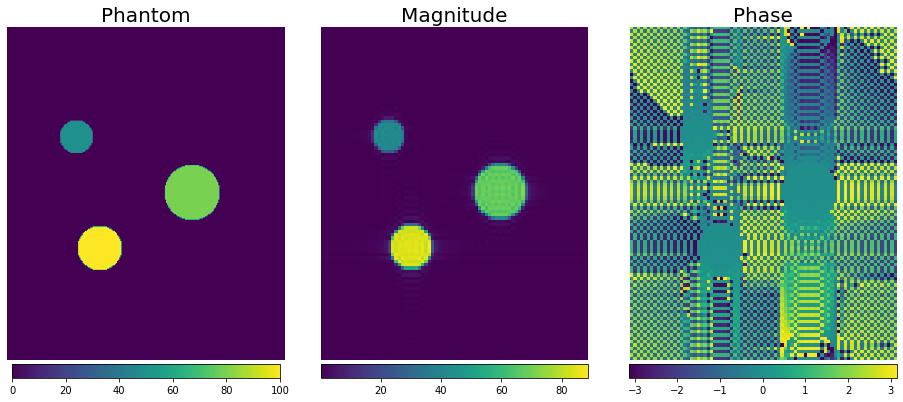
f, a = plt.subplots(1, 3, figsize=(13, 6))
magnitude_image = np.squeeze(np.abs(simulation_result))
phase_image = np.squeeze(np.angle(simulation_result))
img0 = a[0].imshow(phantom_reference_map, origin="lower")
img1 = a[1].imshow(magnitude_image, origin="lower")
img2 = a[2].imshow(phase_image, origin="lower")
[plt.colorbar(im, ax=ax, location="bottom", pad=0.01, shrink=0.9) for im, ax in zip([img0, img1, img2], a)]
[_.axis("off") for _ in a.flatten()]
[_.set_title(t, fontsize=20) for _, t in zip(a, ("Phantom", "Magnitude", "Phase"))]
f.tight_layout()
Auxilliary functions#
tf.train.Checkpoint, and afterwards be restored from the checkpoint by calling the classmethod SimulationBase.from_checkpoint(...). To ensure that the parameters are equal, the simulation is redone and the results are compared.[9]:
import os
# Specify saving path
chkpt = './model_checkpoints/example_checkpoint'
os.makedirs(os.path.dirname(chkpt), exist_ok=True)
# Save simulation to checkpoint and delete old instance
simulator.save(checkpoint_path=chkpt)
del simulator
# construct new intance from checkpoint
new_simulator = ExampleSimulation.from_checkpoint(chkpt)
[10]:
%time simulation_load_result = new_simulator(dataset, voxel_batch_size=5000, execute_eager=True)
print("If this is equal to zero, the simulated results are identical:\n\t Difference of simulated k-space ->",
tf.reduce_sum(simulation_result - simulation_load_result))
Run Simulation: |XXXXXXXXXXXXXXX|3883/3883 |XXXXXXXXXXXXXXX|100/100 CPU times: total: 1.98 s
Wall time: 1.37 s
If this is equal to zero, the simulated results are identical:
Difference of simulated k-space -> tf.Tensor(0j, shape=(), dtype=complex64)
tf.Variable.assign() method. To show if the results differ after setting different Echo-Times the simulated images are compared again:[11]:
new_simulator.forward_model.spin_echo.TE.assign([10,]) # in milliseconds
%time n = new_simulator(dataset, voxel_batch_size=5000)
print('Mean absolute difference for different TE:', tf.reduce_mean(tf.abs(simulation_load_result - n)))
new_simulator.forward_model.spin_echo.TE.assign([50,]) # in milliseconds
%time n = new_simulator(dataset, voxel_batch_size=5000)
print('Mean absolute difference for same TE:', tf.reduce_mean(tf.abs(simulation_load_result - n)))
Run Simulation: |XXXXXXXXXXXXXXX|3883/3883 |XXXXXXXXXXXXXXX|100/100 CPU times: total: 3.97 s
Wall time: 2.37 s
Mean absolute difference for different TE: tf.Tensor(4.6015587, shape=(), dtype=float32)
Run Simulation: |XXXXXXXXXXXXXXX|3883/3883 |XXXXXXXXXXXXXXX|100/100 CPU times: total: 3.16 s
Wall time: 2.38 s
Mean absolute difference for same TE: tf.Tensor(0.0, shape=(), dtype=float32)
[12]:
import json
from IPython.display import JSON
summary_dict = new_simulator.configuration_summary
JSON(json.dumps(summary_dict))
C:\Users\Jonat\anaconda3\envs\tf29_pyvista\lib\site-packages\IPython\core\display.py:606: UserWarning: JSON expects JSONable dict or list, not JSON strings
warnings.warn("JSON expects JSONable dict or list, not JSON strings")
[12]:
<IPython.core.display.JSON object>