Contrast Modules#
All contrast modules are implemented as a subclass to BaseSignalModel. To ensure extensibility stable functionality all modules need to take the same inputarguments and return a tensor of the same shape as the input tensor. To make the required properties provided as input availabe to the __call__ methodduring runtime (while maintaining the uniform calling-argument signature) they have to bepassed via **kwargs (keyword-arguments). To make sure they are loaded during runtime, all required properties have to be specifiedin the ‘required_quantities’ class attribute. A table of the implemented modules alongwith their required input is shown below:
Required Quantities#
Sub - module |
Required Quantities |
|---|---|
BaseSignalModel |
() |
LookUpTableModule |
(‘r_vectors’,) |
CoilSensitivity |
(‘r_vectors’,) |
BirdcageCoilSensitivities |
(‘r_vectors’,) |
GaussianDiffusionTensor |
(‘diffusion_tensor’,) |
PhaseTracking |
(‘trajectory’,) |
SpinEcho |
(‘T1’, ‘T2’) |
SaturationRecoveryGRE |
(‘T1’,) |
BSSFP |
(‘T1’, ‘T2’, ‘T2star’, ‘off_res’) |
StaticT2starDecay |
(‘T2star’,) |
OffResonanceProperty |
(‘deltaB0’,) |
SliceProfile |
(‘r_vectors_excitation’,) |
LocalLookREST |
(‘r_vectors_excitation’,) |
Summary#
All concrete implementations of modules that modify the transverse magnetization need to be sub-classed from this abstract class. |
Base implementation of a signal process that involves a spatial look-up in a T + 3D + channel map. |
Lookup for custom coil sensitivities |
Lookup for ideal bird-cage coil sensitivities. |
Model based Gaussian diffusion contrast. |
Evalulates Phase accrual for pre-defined particle trajectories |
|
Analytical Solution operator for a Single echo Spin echo sequence |
Analytical solution for a Saturation Recovery GRE sequence used in 1st-pass-perfusion imaging |
|
Evaluates bssfp steady state contrast for \(TE/TR \approx 1/2\) according to: ` Ganter, MRM 56:687 (2006)` |
Evaluation of T2-star effect. Initializes a module to calculate the T2* decay |
Initializes a module to calculate the phase contribution of an offresonance according to: |
|
Simplified slice-selection module, computing a weighting from 0 to 1 depending on through-slice position. |
Simplified Local Look with REST slabs module, computing a weighting from 0 to 1 depending on MPS position (Box-selective excitation). |
Modules#
- class BaseSignalModel(expand_repetitions, name=None, device=None)[source]#
All concrete implementations of modules that modify the transverse magnetization need to be sub-classed from this abstract class. All sub-classes must implement the __call__ method
Class to inherit every module from, that multiplies a term Psi to the integrand in the fourier transform:
\[s(\mathbf{k}(t)) = \int_V \mathrm{d}V m( \mathbf{r} , 0) \Psi(\mathbf{r}, t) e^{-2\pi j \mathbf{k}(t) \cdot \mathbf{r}}\]The purpose of this parent class is to ensure the attributes ‘’ required quantities field is set and the __call__ method is implemented
- Parameters:
Methods:
__call__(signal_tensor, **kwargs)Defines the calculation which represent the factor multiplied to the integrand in the signal equation.
check_inputs(**kwargs)This method is meant to be called by CompositeSignalModel and asserts that all required_quantities are passed in kwargs during graph construction.
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
Attributes:
Name of the device that the module is executed on (defaults to: GPU:0 - CPU:0)
- abstract __call__(signal_tensor, **kwargs)[source]#
Defines the calculation which represent the factor multiplied to the integrand in the signal equation. Tensors and arguments that are listed in the self._required_quantities field are automatically passed via kwargs if the module is inserted into a
CompositeSignalModel.- Parameters:
signal_tensor – tf.complex64 - Tensor of points representing local magnetization multiplied with factors of previously executed modules
kwargs – all available tensors representing quantities at given points
- Returns:
signal_tensor * (defined_factor)
- check_inputs(**kwargs)[source]#
This method is meant to be called by CompositeSignalModel and asserts that all required_quantities are passed in kwargs during graph construction.
- Raises:
ValueError if **kwargs is missing a specified required quantity
- abstract update()[source]#
Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it. In other cases, e.g. when the expansion factor changes on Variable change, this needs to be mirrored here.
Function is called before every simulation execute.
- class LookUpTableModule(look_up_map3d, map_dimensions, name=None, expand_repetitions=True, **kwargs)[source]#
Base implementation of a signal process that involves a spatial look-up in a T + 3D + channel map.
- Parameters:
look_up_map3d (
ndarray) – (X, Y, Z, #channels,) where X, Y, Z are the number of bins of the 3D map. Or if temporal information is present: (t, X, Y, Z, #channels,)map_dimensions (
ndarray) – (3, 2) -> [(xlow, xhigh), (ylow, yhigh), (zlow, zhigh)] Extend of the map in scanner coordinate system. Used to sort iso-chromat positions into bins. This is assumed to be the ‘length’ in meter in each spatial direction ofexpand_repetitions (
bool) – See BaseSignalModel for explanationkwargs –
device: str defaults to CPU:0
Methods:
__call__(r_vectors[, method])Evaluates look up of stored values for given positional vectors.
get_indices(r_vectors)Rounds r_vectors to map-resolution to get look-up indices that can be used to gather the corresponding values :type r_vectors: :param r_vectors: () :return:
linear_interpolation(r_vectors[, batch_index])Rounds r_vectors to map-resolution to get look-up indices and gathers the required values from the stored 3D table.
nearest_neighbour(r_vectors[, batch_index])Rounds r_vectors to map-resolution to get look-up indices and gathers the required values from the stored 3D table.
update()If the look-up map is changed, the look-up parameters have to be updated as well
Attributes:
Three dimensional look-up-table assuming a regular spacing derived from self.map-dimension
Spatial extend of the look up as: (3, 2) -> [(xlow, xhigh), (ylow, yhigh), (zlow, zhigh)]
Data type of the lookup map
- __call__(r_vectors, method='nearest', **kwargs)[source]#
Evaluates look up of stored values for given positional vectors.
- Parameters:
r_vectors – (#batch, 3)
method (
str) – one of [“nearest”, “trilinear”] determines which interpolation strategy is used for look-upkwargs – placeholder to allow arbitrary keyword arguments
- Returns:
- get_indices(r_vectors)[source]#
Rounds r_vectors to map-resolution to get look-up indices that can be used to gather the corresponding values :type r_vectors: :param r_vectors: () :return:
- linear_interpolation(r_vectors: ~tensorflow.python.framework.tensor.Tensor, batch_index: ~tensorflow.python.framework.tensor.Tensor = <tf.Tensor: shape=(1,), dtype=int32, numpy=array([0], dtype=int32)>, **kwargs) Tensor[source]#
Rounds r_vectors to map-resolution to get look-up indices and gathers the required values from the stored 3D table.
- Parameters:
r_vectors (
Tensor) – (-1, 3) in meterbatch_index (
Tensor) – (#batch) indices of the first dimension (T) used for spatial lookup
- Return type:
Tensor- Returns:
(#batch_idx, -1, self._n_channels) trilinear interpolated map values for specified locations
- nearest_neighbour(r_vectors: ~tensorflow.python.framework.tensor.Tensor, batch_index: ~tensorflow.python.framework.tensor.Tensor = <tf.Tensor: shape=(1,), dtype=int32, numpy=array([0], dtype=int32)>, **kwargs) Tensor[source]#
Rounds r_vectors to map-resolution to get look-up indices and gathers the required values from the stored 3D table.
- Parameters:
r_vectors (
Tensor) – (-1, 3) in meterbatch_index (
Tensor) – (#batch) indices of the first dimension (T) used for spatial lookup
- Return type:
Tensor- Returns:
(-1, self._n_channels) map values at nearest neighbour locations
-
look_up_map3d:
Variable# Three dimensional look-up-table assuming a regular spacing derived from self.map-dimension
-
map_dimensions:
Variable# Spatial extend of the look up as: (3, 2) -> [(xlow, xhigh), (ylow, yhigh), (zlow, zhigh)]
-
map_dtype:
DType# Data type of the lookup map
- class CoilSensitivity(coil_sensitivities, map_dimensions, device='GPU:0', **kwargs)[source]#
Lookup for custom coil sensitivities
Instantiates a Module that looks up coil sensitivities and multiplies the sensitivy value for the corresponding position to the signal vector when called.
Note
expand_repetitions is always set to True for coil-sensitivities, which means that the output has a #repetions * #coils dimension on the repetitions axis
- Parameters:
coil_sensitivities (
Union[ndarray,Tensor]) – (X, Y, Z, #channels) with dtype complex64map_dimensions (
ndarray) – (3, 2) -> [(xlow, xhigh), (ylow, yhigh), (zlow, zhigh)] Extend of the map in scanner coordinate system. Used to sort iso-chromat positions into bins. This is assumed to be the length in meter in each spatial directiondevice (
str) – (str) Device on which all operations except for the look-up are placed (default: GPU:0)device_lookup – (str) Device where the lookup table is places (defaults to CPU:0)
Methods:
__call__(signal_tensor, r_vectors, **kwargs)Evaluation of coil-sensitivity weighting:
from_3d_birdcage(map_shape, map_dimensions)Lookup for ideal bird-cage coil sensitivities.
lookup_complex_coil_factors(r_vectors)Looks the sensitivity values for all coils at specified locations.
update()If the look-up map is changed, the look-up parameters have to be updated as well
Attributes:
Property returning the complex coil-sensitivities
Three dimensional look-up-table assuming a regular spacing derived from self.map-dimension
Spatial extend of the look up as: (3, 2) -> [(xlow, xhigh), (ylow, yhigh), (zlow, zhigh)]
- __call__(signal_tensor: Tensor, r_vectors: Tensor, **kwargs) Tensor[source]#
Evaluation of coil-sensitivity weighting:
\[M_{xy}^{n}(\vec{r}) = M_{xy}(\vec{r}) * C^{n}(\vec{r})\]- Parameters:
signal_tensor (
Tensor) –r_vectors (
Tensor) – (#voxels, #repetitions, #samples, 3)
- Return type:
Tensor- Returns:
- classmethod from_3d_birdcage(map_shape, map_dimensions, relative_coil_radius=1.5, device='CPU:0', **kwargs)[source]#
Lookup for ideal bird-cage coil sensitivities.
Note
expand_repetitions is always set to True for coil-sensitivities, which means that the output has a #repetions * #coils dimension on the repetitions axis
- Parameters:
map_shape (
Union[ndarray,Tensor,Tuple]) – (X, Y, Z, #coils) array shape of the resulting coil-mapsmap_dimensions (
ndarray) – (3, 2) -> [(xlow, xhigh), (ylow, yhigh), (zlow, zhigh)] Extend ofrelative_coil_radius (
float) – distance of the birdcage ring from the map center the map in scanner coordinate system. Used to sort iso-chromat positions into bins. This is assumed to be the length in meter in each spatial directiondevice (
str) – (str) Device on which all operations except for the look-up are placed (default: GPU:0)device_lookup – (str) Device where the lookup table is places (defaults to CPU:0)
- lookup_complex_coil_factors(r_vectors)[source]#
Looks the sensitivity values for all coils at specified locations.
- Parameters:
r_vectors (
Tensor) – […, 3] dtype: tf.float32- Returns:
coil_factors […, n_coils] dtype: tf.complex64
- property coil_maps#
Property returning the complex coil-sensitivities
-
look_up_map3d:
Variable# Three dimensional look-up-table assuming a regular spacing derived from self.map-dimension
-
map_dimensions:
Variable# Spatial extend of the look up as: (3, 2) -> [(xlow, xhigh), (ylow, yhigh), (zlow, zhigh)]
-
map_dtype:
DType# Data type of the lookup map
- class BirdcageCoilSensitivities(map_shape, map_dimensions, relative_coil_radius=1.5, device='CPU:0', **kwargs)[source]#
Lookup for ideal bird-cage coil sensitivities.
Instantiates a Module that looks up coil sensitivities and multiplies the sensitivity value for the corresponding position to the signal vector when called. The coil sensitivities are constructed as ideal birdcage-sensitivities.
Note
expand_repetitions is always set to True for coil-sensitivities, which means that the output hast a #repetions * #coils dimension on the repetitions axis
- Parameters:
map_shape (
Union[ndarray,Tensor,Tuple]) – (X, Y, Z, #coils) array shape of the resulting coil-mapsrelative_coil_radius (
float) – distance of the birdcage ring from the map centermap_dimensions (
ndarray) – (3, 2) -> [(xlow, xhigh), (ylow, yhigh), (zlow, zhigh)] Extend of the map in scanner coordinate system. Used to sort iso-chromat positions into bins. This is assumed to be the length in meter in each spatial directiondevice (
str) – (str) Device on which all operations except for the look-up are placed (default: GPU:0)device_lookup – (str) Device where the lookup table is places (defaults to CPU:0)
- class GaussianDiffusionTensor(b_vectors, expand_repetitions, **kwargs)[source]#
Model based Gaussian diffusion contrast.
Instantiates a module that evaluates the diffusion signal representation:
\[F = \exp(-\mathbf{b}^T\mathbf{D}\mathbf{b})\]Assumes the b-vector to be the diffusion direction be scaled with the square-root of the b-value: \(\mathbf{b} = \sqrt{b}\mathbf{d}\)
Instead of two dimensions for direction and b-value just uses one dimension of scaled b-vectors.
- Parameters:
b_vectors (
Tensor) –(#directions * #bvalues, 3) in units sqrt(s/mm*2)
expand_repetitions (
bool) – See BaseSignalModel for explanation
Methods:
__call__(signal_tensor, diffusion_tensor, ...)Evaluates the diffusion operator.
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
- __call__(signal_tensor: Tensor, diffusion_tensor: Tensor, **kwargs) Tensor[source]#
Evaluates the diffusion operator.
- Parameters:
signal_tensor (
Tensor) – (#voxel, #repetitions, #k-space-samples) last two dimensions can be 1diffusion_tensor (
Tensor) – diffusion_tensor (#voxels, #repetitions, #k_space_samples, 3, 3)
- Return type:
Tensor- Returns:
- if expand_repetitions == True: (#voxel, #repetitions * #self.b_vectors,
#k-space-samples)
if expand_repetitions == False: (#voxel, #repetitions, #k-space-samples)
- class PhaseTracking(expand_repetitions, wave_form, gamma, **kwargs)[source]#
Evalulates Phase accrual for pre-defined particle trajectories
Phase calculation for a simple spatially constant gradient wave form and for moving particles according to:
\[\phi(t) = \int \vec{G}(t) \cdot \vec{r}(t) dt\]- Parameters:
wave_form (
Tensor) – (#repetition, #steps, [t, x, y, z]) - time and wave-form-gridgamma (
float) – Gyromagnetic ratio in rad/ms/mTexpand_repetitions (Variable) –
Methods:
__call__(signal_tensor, trajectory, **kwargs)Evaluates the phase integration for given trajectories
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
Attributes:
gyromagnetic ratio in rad/mT/ms
(#repetitions, #time-steps, 4[t, x, y, z])
- __call__(signal_tensor, trajectory, **kwargs)[source]#
Evaluates the phase integration for given trajectories
- Parameters:
signal_tensor (
Tensor) – (#voxel, #repetitions, #k-space-samples) last two dimensions can be 1trajectory (
Tensor) – (#voxel, #repetitions, #k-space-samples, #time-steps, 3) last dimension corresponds to (x, y, z) in meter #time-steps must be the same as self.wave_form
- Return type:
Tensor- Returns:
- if expand_repetitions == True: (#voxel, #repetitions * self.wave_form.shape[0],
#k-space-samples)
if expand_repetitions == False: (#voxel, #repetitions, #k-space-samples)
- update()[source]#
Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it. In other cases, e.g. when the expansion factor changes on Variable change, this needs to be mirrored here.
Function is called before every simulation execute.
-
gamma:
Tensor# gyromagnetic ratio in rad/mT/ms
-
wave_form:
Variable# (#repetitions, #time-steps, 4[t, x, y, z])
- class SpinEcho(echo_time, repetition_time, expand_repetitions, **kwargs)[source]#
Analytical Solution operator for a Single echo Spin echo sequence
Evaluates the signal for the simple Spin-Echo sequence:
\[M_{xy} = M_z \cdot e^{- TE / T_2} (1 - e^{-TR / T1})\]- Raises:
ValueError if the arguments echo and repetition time do not have the same length
- Parameters:
Methods:
__call__(signal_tensor, T1, T2, **kwargs)Evaluates spin echo operator
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
Attributes:
Echo time in ms
Repetition time in ms
- __call__(signal_tensor: Tensor, T1: Tensor, T2: Tensor, **kwargs)[source]#
Evaluates spin echo operator
- Parameters:
signal_tensor (
Tensor) – (#batch, #voxel, #repetitions, #k-space-samples) last two dimensions can be 1T1 (
Tensor) – (#batch, #voxel, 1, 1) in millisecondsT2 (
Tensor) – (#batch, #voxel, 1, 1) in milliseconds
- Returns:
- update()[source]#
Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it. In other cases, e.g. when the expansion factor changes on Variable change, this needs to be mirrored here.
Function is called before every simulation execute.
-
TE:
Variable= None# Echo time in ms
-
TR:
Variable= None# Repetition time in ms
- class SaturationRecoveryGRE(repetition_time, flip_angle, saturation_delay, n_profiles_to_k0, expand_repetitions, **kwargs)[source]#
Analytical solution for a Saturation Recovery GRE sequence used in 1st-pass-perfusion imaging
Evaluates the signal model for a saturation recovery spoiled gradient echo sequence. This modules assumes the cartesian line-by line k-space sampling. This yields the signal equation for a single k-space line:
\[m_{xy} = \rho(r) \left[(1 - e^{-T_D \cdot R_1}) (\cos(\alpha)e^{-T_R \cdot R_1})^{n-1} + (1-e^{-T_R \cdot R_1}) \frac{1 - (\cos(\alpha)e^{-T_R \cdot R_1})^{n-1}} {1 - (\cos(\alpha)e^{-T_R \cdot R_1})} \right]\]Where T_D is the saturation_delay, n is the number of profiles left until the k-space-center, alpha is the flip_angle and TR is the repetition time.
for more info refer to https://gitlab.ethz.ch/jweine/cmrsim/-/issues/62
Note This module assumes, that the keyword argument ‘segment_index’ is used in __call__`, such that each segment contains all k-space point acquired in one TR, otherwise the signal equation does not hold!
- Parameters:
Methods:
__call__(signal_tensor, T1[, segment_index])Evaluation of SaturationRecoveryGRE equation
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
Attributes:
Echo time in ms
Repetition time in ms
- __call__(signal_tensor, T1, segment_index=0.0, **kwargs)[source]#
Evaluation of SaturationRecoveryGRE equation
- Parameters:
signal_tensor (Tensor) –
T1 (Tensor) –
segment_index (Tensor) –
- update()[source]#
Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it. In other cases, e.g. when the expansion factor changes on Variable change, this needs to be mirrored here.
Function is called before every simulation execute.
-
TE:
Variable= None# Echo time in ms
-
TR:
Variable= None# Repetition time in ms
- class BSSFP(flip_angle, echo_time, repetition_time, expand_repetitions, **kwargs)[source]#
Evaluates bssfp steady state contrast for \(TE/TR \approx 1/2\) according to: ` Ganter, MRM 56:687 (2006)`
\[M(x \cdot TR) = E_2^x e^{ix\theta} \left[ (E_2^{\prime})^x [1 + e^{i\theta} E_2^{\prime} A]^{-1} + (E_2^{\prime})^{1-x} e^{-i\theta} \Lambda/E_2 [1 + e^{-i\theta} E_2^{\prime} A]^{-1} \right]\]Off-resonance theta results in banding according to the alternating RF phase scheme:
Off-resonance Banding
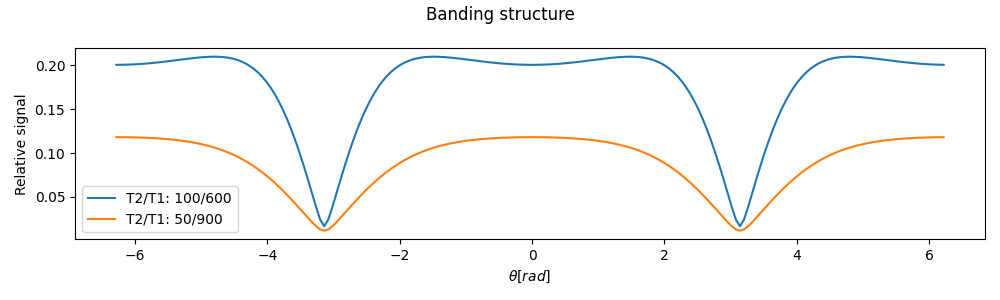
- Raises:
ValueError if the arguments echo, repetition time and flip angle do not have the same length
- Parameters:
Methods:
__call__(signal_tensor, T1, T2, T2star, ...)Evaluates the signal Equation.
Attributes:
Flip angle in degree
Echo time in ms
Repetition time in ms
- __call__(signal_tensor, T1, T2, T2star, off_res, **kwargs)[source]#
Evaluates the signal Equation.
- Parameters:
signal_tensor (
Tensor) – (#voxel, #repetitions, #k-space-samples) last two dimensions can be 1T1 (
Tensor) – in msT2 (
Tensor) – in msT2star (
Tensor) – in msoff_res (
Tensor) – b0 inhomogeneity in rad/ms (angular frequency difference)
- Returns:
(#voxel, #repetitions, #k-space-samples)
-
FA:
Variable= None# Flip angle in degree
-
TE:
Variable= None# Echo time in ms
-
TR:
Variable= None# Repetition time in ms
- class StaticT2starDecay(sampling_times, expand_repetitions, **kwargs)[source]#
- Evaluation of T2-star effect. Initializes a module to calculate the T2* decay
sampling times in milliseconds, according to:
\[M_{xy}^{i, \prime} = M_{xy}^{i} exp(-\delta/T2*),\]where \(\delta\) is the duration from the RF-excitation center for GRE sequences while it is the temporal distance to the echo-center for SE sequences.
If sequence is Spin-Echo, the sampling times before echo-center have to be negative.
- Parameters:
Methods:
__call__(signal_tensor, T2star, **kwargs)Evaluates the T2* operator
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
- __call__(signal_tensor: Tensor, T2star: Tensor, **kwargs)[source]#
Evaluates the T2* operator
- Parameters:
signal_tensor (
Tensor) – (#voxel, #repetitions, #k-space-samples) #repetitions and #k-space-samples can be1T2star (
Tensor) – tf.Tensor (#voxel, 1, 1) in millisecondssegment_index – int, if k-space is segmented for Memory reasons. defaults to 0
- Returns:
signal_tensor (#voxel, #repetition, #k-space-samples) with #k-space-samples > 1
- class OffResonanceProperty(sampling_times, gamma, expand_repetitions, **kwargs)[source]#
Initializes a module to calculate the phase contribution of an offresonance according to:
\[M_{xy}^{i, \prime} = M_{xy}^{i} exp(-j\gamma \Delta B \delta t),\]where \(\delta t\) is the duration from the RF-excitation center for GRE sequences while it is the temporal distance to the echo-center for SE sequences.
Note
This module does not expand the repetition axis, but it always broad-casts the internal calculations to work for all #repetions > 0
- Parameters:
Methods:
__call__(signal_tensor, deltaB0, **kwargs)Evaluates the off-resonance operator under the assumption
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
Attributes:
Tuple of names specifying the input quantities that must be passed as keyword-arguments to the __call__ method of the operator instance.
timing of acquisition (ADC) events in milliseconds Assumes the duration from the RF-excitation center for GRE sequences and the temporal distance to the echo-center for SE sequences.
- __call__(signal_tensor: Tensor, deltaB0: Tensor, **kwargs)[source]#
Evaluates the off-resonance operator under the assumption
- Parameters:
signal_tensor (
Tensor) – (#voxel, #repetitions, #k-space-samples) #repetitions and #k-space-samples can be1deltaB0 (
Tensor) – tf.Tensor (#voxel, 1, 1) in mT.segment_index – int, if k-space is segmented for Memory reasons. defaults to 0
- Returns:
signal_tensor (#voxel, #repetition, #k-space-samples) with #k-space-samples > 1
- update()[source]#
Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it. In other cases, e.g. when the expansion factor changes on Variable change, this needs to be mirrored here.
Function is called before every simulation execute.
- required_quantities: Tuple[str] = ('deltaB0',)#
Tuple of names specifying the input quantities that must be passed as keyword-arguments to the __call__ method of the operator instance.
-
sampling_times:
Variable# timing of acquisition (ADC) events in milliseconds Assumes the duration from the RF-excitation center for GRE sequences and the temporal distance to the echo-center for SE sequences.
- class SliceProfile(expand_repetitions, slice_normal, slice_position, slice_thickness, device=None)[source]#
Simplified slice-selection module, computing a weighting from 0 to 1 depending on through-slice position.
Note
To modify the actual slice-profile you can implement a subclass of this class that implements a different ‘slice_profile’ method.
Example Images

Example Usage
uniform_mesh = pyvista.UniformGrid(spacing=(0.001, 0.001, 0.001), dimensions=(100, 100, 100), origin=(-0.05, -0.05, -0.05)) r_vectors = tf.constant(uniform_mesh.points, dtype=tf.float32) slice_normal = np.array([[0, 1, 1], [0, 0, 1], [1, 2, 0]], dtype=np.float64) slice_normal /= np.linalg.norm(slice_normal, keepdims=True, axis=-1) slice_position = Quantity([[0, 1, 1], [1, 0, 0], [0, 1, 0]], "cm").m_as("m") slice_thickness = Quantity([2, 4, 0.5], "cm").m_as("m") slice_mod = cmrsim.analytic.contrast.SliceProfile(expand_repetitions=True, slice_normal=slice_normal, slice_position=slice_position, slice_thickness=slice_thickness) r_vectors_excitation = tf.reshape(r_vectors, [-1, 1, 1, 3]) signal_start = tf.ones(r_vectors_excitation.shape[:-1], dtype=tf.complex64) signal_out = slice_mod(signal_tensor=signal_start, r_vectors_excitation=r_vectors_excitation).numpy() for i in range(signal_out.shape[1]): uniform_mesh[f"signal_out{i}"] = np.abs(signal_out)[:, i, 0]
- Parameters:
expand_repetitions (
bool) – if True, expands repetition axes with the factor determined by the first axes of the following input argumentsslice_normal (
Sequence[Union[float,Sequence[float]]]) – (expansion_factor, 3) vector defining the slice normal of the excitation sliceslice_position (
Sequence[Union[float,Sequence[float]]]) – (expansion_factor, 3) vector determining the slice-center-pointdevice (str) –
:param slice_thickness:(expansion_factor, ) scalar in meter, determining the slice thickness :type device:
str:param device:Methods:
__call__(signal_tensor, ...)Call function for analytice slice-profile weighting
slice_profile(s_coord)- type s_coord:
Tensor
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
Attributes:
Additional mandatory keyword arguments for call
(expansion_factor, 3) vector defining the slice normal of the excitation slice
(expansion_factor, 3) vector determining the slice-center-point
(expansion_factor, ) scalar in meter, determining the slice thickness
- __call__(signal_tensor, r_vectors_excitation, **kwargs)[source]#
Call function for analytice slice-profile weighting
- Raises:
AssertionError - r_vectors_excitation.shape[1] not equal to 1 or self.expansion_factor - r_vectors_excitation.shape[2] is not equal to 1 (k-samples not supported here)
- Parameters:
signal_tensor (
Tensor) – (#batch, [#repetitions, 1], #ksamples)r_vectors_excitation (
Tensor) – (#batch, [#repetition, 1], 1, 3)
- Returns:
signal_tensor weighted by slice selective excitation
- slice_profile(s_coord)[source]#
- Parameters:
s_coord (
Tensor) – arbitrary shaped tensor containing the values for through-slice coordinate relative to slice position- Return type:
Tensor- Returns:
factor between 0, 1 for each position (of stame shape as s_coord)
- update()[source]#
Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it. In other cases, e.g. when the expansion factor changes on Variable change, this needs to be mirrored here.
Function is called before every simulation execute.
-
required_quantities:
Tuple[str] = ('r_vectors_excitation',)# Additional mandatory keyword arguments for call
-
slice_normal:
Variable= None# (expansion_factor, 3) vector defining the slice normal of the excitation slice
-
slice_position:
Variable= None# (expansion_factor, 3) vector determining the slice-center-point
-
slice_thickness:
Variable= None# (expansion_factor, ) scalar in meter, determining the slice thickness
- class LocalLookREST(expand_repetitions, slice_normal, readout_direction, phase_encoding_direction, slice_position, spatial_extends, device=None)[source]#
Simplified Local Look with REST slabs module, computing a weighting from 0 to 1 depending on MPS position (Box-selective excitation).
Note
- To modify the actual profile per M/P/S direction you can create a subclass of this class that
implements a different ‘box_profile’ method.
Example Images

Example Usage
uniform_mesh = pyvista.UniformGrid(spacing=(0.001, 0.001, 0.001), dimensions=(100, 100, 100), origin=(-0.05, -0.05, -0.05)) r_vectors = tf.constant(uniform_mesh.points, dtype=tf.float32) slice_normal = np.eye(3, 3) readouts = np.roll(np.eye(3, 3), 1, axis=0) phase_encodes = np.roll(np.eye(3, 3), 2, axis=0) slice_position = Quantity([[0, 1, 1], [1, 0, 0], [0, 0, 1]], “cm”).m_as(“m”) spatial_extends = Quantity([[3, 2, 0.5], [5, 10, 1], [8, 6, 2]], “cm”).m_as(“m”) * 1.5
rotation_matrices = tf.stack([readouts, phase_encodes, slice_normal], axis=1)
- lolo_mod = cmrsim.analytic.contrast.LocalLookREST(expand_repetitions=True, slice_normal=slice_normal,
readout_direction=readouts, phase_encoding_direction=phase_encodes, slice_position=slice_position, spatial_extends=spatial_extends)
r_vectors_excitation = tf.reshape(r_vectors, [-1, 1, 1, 3]) signal_start = tf.ones(r_vectors_excitation.shape[:-1], dtype=tf.complex64)
signal_out = lolo_mod(signal_tensor=signal_start, r_vectors_excitation=r_vectors_excitation).numpy() for i in range(signal_out.shape[1]):
uniform_mesh[f”signal_out{i}”] = np.abs(signal_out)[:, i, 0]
- Parameters:
expand_repetitions (
bool) – if True, expands repetition axes with the factor determined by the first axes of the following input argumentsslice_normal (
Sequence[Union[float,Sequence[float]]]) – (expansion_factor, 3) vector defining the slice normal of the excitation slicereadout_direction (
Sequence[Union[float,Sequence[float]]]) – (expansion_factor, 3) vector defining the readout directionphase_encoding_direction (
Sequence[Union[float,Sequence[float]]]) – (expansion_factor, 3) vector defining the phase encoding directionslice_position (
Sequence[Union[float,Sequence[float]]]) – (expansion_factor, 3) vector determining the slice-center-pointspatial_extends (
Sequence[Union[float,Sequence[float]]]) – (expansion_factor, 3) box-width per M-P-S direction around the slice-positiondevice (
str) –
Methods:
__call__(signal_tensor, ...)Call function for analytice local look with REST slabs weighting
box_profile(mps_coords)- type mps_coords:
Tensor
update()Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it.
Attributes:
(expansion_factor, 4, 4) orientation matrix containing the transformation matrix from r_vectors into MPS coordinates.
Additional mandatory keyword arguments for call
(expansion_factor, 3) spatial extend per M-P-S directions (conceptually equivalent to slice-thickness)
- __call__(signal_tensor, r_vectors_excitation, **kwargs)[source]#
Call function for analytice local look with REST slabs weighting
- Raises:
AssertionError - r_vectors_excitation.shape[1] not equal to 1 or self.expansion_factor - r_vectors_excitation.shape[2] is not equal to 1 (k-samples not supported here)
- Parameters:
signal_tensor (
Tensor) – (#batch, [#repetitions, 1], #ksamples)r_vectors_excitation (
Tensor) – (#batch, [#repetition, 1], 1, 3)
- Returns:
signal_tensor weighted by box-selective excitation
- box_profile(mps_coords)[source]#
- Parameters:
mps_coords (
Tensor) – arbitrary shaped tensor containing the values for MPS coordinate relative to slice position- Return type:
Tensor- Returns:
factor between 0, 1 for each position (of same shape as mps_coords[…, 0])
- update()[source]#
Can be left empty if changing the Variables in the signal module does not affect the shape of the tensors processed by it. In other cases, e.g. when the expansion factor changes on Variable change, this needs to be mirrored here.
Function is called before every simulation execute.
-
orientation_matrix:
Variable# (expansion_factor, 4, 4) orientation matrix containing the transformation matrix from r_vectors into MPS coordinates. Is composed from slice_normal, phase_encoding and readout directions. matrix per repetition is guaranteed to be orthogonal
-
required_quantities:
Tuple[str] = ('r_vectors_excitation',)# Additional mandatory keyword arguments for call
-
spatial_extend:
Variable# (expansion_factor, 3) spatial extend per M-P-S directions (conceptually equivalent to slice-thickness)